Research Article |
|
Corresponding author: J. Jennifer Luangsa-ard ( jajen@biotec.or.th ) Corresponding author: Anto Budiharjo ( anto.budiharjo@live.undip.ac.id ) Academic editor: Thorsten Lumbsch
© 2020 Arifah Nur Aini, Suchada Mongkolsamrit, Wijanarka Wijanarka, Donnaya Thanakitpipattana, J. Jennifer Luangsa-ard, Anto Budiharjo.
This is an open access article distributed under the terms of the Creative Commons Attribution License (CC BY 4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Citation:
Aini AN, Mongkolsamrit S, Wijanarka W, Thanakitpipattana D, Luangsa-ard JJ, Budiharjo A (2020) Diversity of Akanthomyces on moths (Lepidoptera) in Thailand. MycoKeys 71: 1-22. https://doi.org/10.3897/mycokeys.71.55126
|
Abstract
Akanthomyces is a genus of invertebrate-pathogenic fungi from the family Cordycipitaceae (Ascomycota, Hypocreales). Its species occurs on two different types of hosts, spiders and insects, and in the latter case specifically Lepidoptera adults. Three new species of Akanthomyces, A. noctuidarum, A. pyralidarum, and A. tortricidarum occurring on adult moths from Thailand are proposed based on the differences of their morphological characteristics and molecular data. Phylogenetic analyses using a combined dataset, including the internal transcribed spacer regions, the large subunit of the ribosomal DNA, translation elongation factor 1-α, the largest subunit of RNA polymerase II, and the second largest subunit of RNA polymerase II, support the delimitation of these new species in Akanthomyces.
Keywords
Akanthomyces, entomopathogenic fungi, fungal taxonomy, multilocus phylogeny
Introduction
Cordycipitaceae is one of the families of the order Hypocreales with entomogenous nutritional habit. Many of the species in this family have been originally isolated from dead insects and spiders that are buried in the soil, leaf litter, or attached to the undersides or upper sides of a leaf. Some species, especially in Beauveria, could be found in the soil (
Akanthomyces was established by
Species complexes or cryptic species are common in the kingdom Fungi. Given the simplicity of the phenotypic characters and the overlap of the size and shapes of important diagnostic features, species in many genera cannot be easily classified and identified. Cryptic species refers to taxa that are morphologically similar, yet evidence has shown that they are on different evolutionary paths as revealed by molecular phylogenetic methods and can only be recognized by their DNA sequences. Entomopathogenic fungi from Thailand are commonly encountered in the forests and constitute a huge number in our collections (
In surveys of entomopathogenic fungi in national parks and community forests, collections of pathogens on adult moths were found on the underside of leaves of dicotyledonous forest plants. The phenotypic characters of the collections in having cylindrical to narrowly clavate synnemata and superficial perithecia scattered on the body and wings of the moth identify them primarily to be members of Akanthomyces in Cordycipitaceae, mostly as Akanthomyces cf. tuberculatus. The aims of this study were (1) to elucidate the relationships of these collections to known members of Cordycipitaceae, (2) to uncover hidden species in A. tuberculatus species complex, and (3) to describe new taxa to accommodate species diversity in Akanthomyces.
Materials and methods
Fungal materials and isolation
The specimens used in this study were obtained from BIOTEC Culture Collection (BCC) and BIOTEC Bangkok Herbarium (BBH), Thailand. Fungal specimens were collected from several national parks in Thailand. Soil from the forest floor, leaf litter, undersides, and upper sides of the leaves were scanned for fungal growth on dead insects. Collected specimens were stored in plastic boxes, returned to the laboratory, and examined under a stereo microscope (Olympus SZ61). Isolation from the teleomorphs followed the method described by
Isolation from the anamorphs was carried out using a sterilized inoculation needle to pick the conidia out from sporulating structures and then transfer them on to a PDA plate. These plates were stored in a plastic box chamber at room temperature, left overnight until the conidia germinated, and treated the same way as described in
Colony growth and morphology
Fungal structures of both, anamorph and teleomorph, such as perithecia, asci, ascospores, synnemata, phialides, and conidia were mounted on glass slides with a drop of lactophenol cotton blue solution. Microscopic measurements of 50 individual fungal structures were obtained using a light microscope (Olympus CX31). Variability was provided as the mean ± standard deviation with absolute minima and maxima in parentheses. Detailed colony descriptions and morphological comparisons of some fungal structures were determined from cultures grown on PDA and OA for 14 days at 25 °C (
DNA extraction
Genomic DNA was extracted from fungal cultures on PDA using a modified CTAB method (
PCR amplification and sequencing
Five nuclear loci regions, namely, internal transcribed spacers 1 and 2 along with the 5.8S rDNA (ITS), large subunit of the ribosomal DNA (LSU), translation elongation factor 1-α (TEF), the largest subunit of RNA polymerase II (RPB1), and the second largest subunit of RNA polymerase II (RPB2), were amplified and sequenced. PCR amplifications were conducted in a 25 µL volume consisting of 1× PCR buffer, 0.4 M betaine, 200 µM of each of the four dNTPs, 1 U Taq DNA polymerase (Thermo Scientific, USA), and 0.2 µM of each primer. The primer pairs used in this study were ITS5 and ITS 4 for ITS (
Sequence alignment and phylogenetic analysis
Each DNA sequence was checked for ambiguous bases and assembled in BioEdit v.7.0.5.3 (
MP analysis used PAUP4.0a116 (
Results
Multilocus phylogeny
A total of 55 new sequences from 11 specimens were obtained in this study (Table 1). ITS sequences were used in a preliminary study to select 11 specimens that represent new species. The combined dataset included 101 taxa and four loci consisting of 3511 bp (LSU 850 bp, TEF 1041 bp, RPB1 732 bp, and RPB2 888 bp). Purpureocillium lilacinum in Ophiocordycipitaceae was used as the outgroup for this dataset.
List of species and GenBank accession numbers of sequences used in this study.
| Species | Strain | Host | GenBank accession numbers | ||||
|---|---|---|---|---|---|---|---|
| ITS | LSU | TEF | RPB1 | RPB2 | |||
| Akanthomyces aculeatus | HUA186145 | – | – | MF416520 f | MF416465 f | – | – |
| TS772 | Lepidoptera; Sphingidae | KC519371 g | KC519370 g | KC519366 g | – | – | |
| Akanthomyces araneogenum | GZUIFDX2 T | Araneus sp. | KU893153 j | MH978179 j | MH978187 j | MH978182 j | MH978185 j |
| GZUIFDX1 | Araneus sp. | KU893152 j | MH978178 j | – | MH978181 j | MH978184 j | |
| GZUIFSN1 | Araneus sp. | MH978177 j | MH978180 j | MH978188 j | MH978183 j | MH978186 j | |
| Akanthomyces attenuatus | CBS402.78 | Leaf litter; Acer saccharum | AJ292434 f | AF339565 f | EF468782 f | EF468888 f | EF468935 f |
| Akanthomyces coccidioperitheciata | NHJ6709 | Araneae; spider | JN049865 f | EU369042 f | EU369025 f | EU369067 f | EU369086 f |
| Akanthomyces farinosa | CBS541.81 | – | – | AY6241807 b | – | JQ4256867b | – |
| Akanthomyces kanyawimiae | TBRC7242 | Araneae; spider | MF140751 i | MF140718 i | MF140838 i | MF140784 i | MF140808 i |
| TBRC7243 | Unidentified | MF140750 i | MF140717 i | MF140837 i | MF140783 i | MF140807 i | |
| Akanthomyces lecanii | CBS101247 | Hemiptera; Coccus viridis | JN049836 f | AF339555 f | DQ522359 f | DQ522407 f | DQ522466 f |
| Akanthomyces muscarius | CBS470.73 | – | – | MH878385 k | – | – | – |
| CBS455.70B | – | – | MH871560 k | – | – | – | |
| CBS455.70C | – | – | MH871561 k | – | – | – | |
| Akanthomyces noctuidarum | BCC36265 T | Lepidoptera; Noctuidae | MT356072 | MT356084 | MT477978 | MT477994 | MT477987 |
| BBH16595 | MT356073 | MT356085 | MT477979 | MT477995 | MT478005 | ||
| BCC47498 | MT356074 | MT356086 | MT477980 | MT477996 | MT477988 | ||
| BCC28571 | MT356075 | MT356087 | MT477981 | MT478009 | MT478006 | ||
| Akanthomyces pyralidarum | BCC28816 T | Lepidoptera; Pyralidae | MT356080 | MT356091 | MT477982 | MT478000 | MT478007 |
| BCC32191 | MT356081 | MT356092 | MT477983 | MT478001 | MT477989 | ||
| BCC40869 | MT356082 | MT356093 | MT477984 | MT478002 | MT477990 | ||
| BCC29197 | MT356083 | MT305694 | MT508840 | MT478003 | MT477991 | ||
| Akanthomyces sulphureus | TBRC7248 T | Araneae; spider | MF140758 i | MF140722 i | MF140843 i | MF140787 i | MF140812 i |
| TBRC7249 | Araneae; spider | MF140757 i | MF140721 i | MF140842 i | MF140786 i | MF140734 i | |
| Akanthomyces thailandicus | TBRC7245 T | Araneae; spider | MF140754 i | – | MF140839 i | – | MF140809 i |
| Akanthomyces tortricidarum | BCC72638 T | Lepidoptera; Tortricidae | MT356076 | MT356088 | MT478004 | MT477997 | MT477992 |
| BCC41868 | MT356077 | MT356089 | MT477985 | MT477998 | MT478008 | ||
| BCC28583 | MT356079 | MT356090 | MT477986 | MT477999 | MT477993 | ||
| Akanthomyces tuberculatus | HUA186131 | Lepidoptera (adult moth) | – | MF416521 h | MF416466 h | – | – |
| Akanthomyces waltergamsii | TBRC7250 | Araneae; spider | MF140749 i | MF140715 i | MF140835 i | – | – |
| TBRC7251 | Araneae; spider | MF140747 i | MF140713 i | MF140833 i | MF140781 i | MF140805 i | |
| Ascopolyporus polychrous | PC 546 | Plant | – | DQ118737 a | DQ118745 a | DQ127236 a | – |
| Ascopolyporus villosus | ARSEF6355 | Plant | – | AY886544 a | DQ118750 a | DQ127241 a | – |
| Beauveria acridophilla | HUA179221 | – | – | JQ895537 g | JQ958615 g | JX003853 g | JX003843 g |
| MCA1181 | Romaleidae; Tropidacris cristata | JQ958607 g | JQ895542 g | – | JX003856 g | – | |
| Beauveria bassiana | ARSEF1564 | Lepidoptera; Arctiidae | HQ880761 e | – | HQ880974 e | HQ880833 e | HQ880905 e |
| Beauveria blattidicola | MCA1727 | – | – | MF416539 h | MF416483 h | MF416640 h | – |
| MCA1814 | – | – | MF416540 h | MF416484 h | MF416641 h | – | |
| Beauveria brongniartii | BCC16585 | Coleoptera; Anomala cuprea (larva) | JN049867 f | JF415967 f | JF4160092 f | JN049885 f | JF415991 f |
| ARSEF617 | Coleoptera; Scarabaeidae | HQ880782 e | – | HQ880991 e | HQ880854 e | HQ880926 e | |
| Beauveria caledonica | ARSEF2567 | Soil | HQ880817 e | AF339520 g | EF469057 g | EF469086 g | HQ880961 g |
| Beauveria malawiensis | ARSEF7760 | Coleoptera; Cerambycidae | – | – | DQ376246 g | HQ880897 g | HQ880969 g |
| Beauveria pseudobassiana | ARSEF3405 | Lepidoptera: Tortricidae | AY532022 e | – | AY531931 e | HQ880864 e | HQ880936 e |
| Blackwellomyces cardinalis | OSC93609 | Lepidoptera; Tineidae (larva) | – | AY184962 g | DQ522325 g | DQ522370 g | DQ522422 g |
| OSC93610 | JN049843 f | AY184963 f | EF469059 f | EF469088 f | EF469106 f | ||
| Cordyceps amoenerosea | CBS107.73 | Coleoptera (pupa) | AY624168 b | MG665224 i | – | – | MG665234 i |
| CBS729.73 | Coleoptera; Nitidulidae | AY624169 b | MG665225 i | HM161732 i | – | MG665235 i | |
| Cordyceps bifusispora | spat08.129 | – | – | MF416523 h | MF416468 h | MF416630 h | – |
| spat08.133.1 | – | – | MF416524 h | MF416469 h | MF416631 h | MF416434 h | |
| Cordyceps blackwelliae | TBRC7253 | Lepidoptera | MF140739 i | MF140705 i | MF140825 i | MF140774 i | MF140798 i |
| TBRC7254 | Lepidoptera | MF140738 i | MF140704 i | MF140824 i | MF140773 i | MF140797 i | |
| TBRC7255 | Lepidoptera | MF140737 i | MF140703 i | MF140823 i | MF140772 i | MF140796 i | |
| Cordyceps caloceroides | MCA2249 | – | – | MF416525 h | MF416470 h | MF416632 h | – |
| QCNE186715 | – | – | MF416526 h | – | – | – | |
| Cordyceps cateniannulata | TBRC7258 | Araneae; spider | MF140753 i | MF140729 i | MF140850 i | MF140767 i | – |
| Cordyceps coleopterorum | CBS110.73 | Coleoptera (larva) | AY624177 f | JF415988 f | JF416028 f | JN049903 f | JF416006 f |
| Cordyceps farinosa | CBS111113 | – | AY624181 b | FJ765253 i | GQ250022 i | – | GU979973 i |
| Cordyceps fumosorosea | CBS375.70 | Food | AY624183 b | MG665229 i | HM161736 i | – | MG665238 i |
| CBS107.10 | – | AY624184 b | MG665227 i | HM161735 i | – | MG665237 i | |
| Cordyceps javanica | TBRC7259 | Lepidoptera | MF140745 i | MF140711 i | MF140831 i | MF140780 i | MF140804 i |
| TBRC7260 | Lepidoptera | MF140744 i | MF140710 i | MF140830 i | MF140779 i | MF140803 i | |
| Cordyceps kyusyuensis | EFCC5886 | Lepidoptera (pupa) | – | EF468813 c | EF468754 c | EF468863 c | EF468917 c |
| Cordyceps lepidopterorum | TBRC7263 | Lepidoptera (larva) | MF140765 i | MF140699 i | MF140819 i | MF140768 i | MF140792 i |
| TBRC7264 | MF140766 i | MF140700 i | MF140820 i | MF140769 i | MF140793 i | ||
| Cordyceps militaris | OSC93623 | Lepidoptera (pupa) | – | EF468821 h | EF468762 h | EF468869 h | – |
| Cordyceps ochraceostromata | ARSEF5691 | Lepidoptera | – | EF468819 h | EF468759 h | EF468867 h | EF468921 h |
| Cordyceps ninchukispora | spat08.115 | – | – | MF416532 h | MF416476 h | MF416635 h | MF416439 h |
| spat09.021 | – | – | MF416533 h | MF416477 h | MF416636 h | – | |
| Cordyceps rosea | spat09.053 | – | – | MF416536 h | MF416480 h | MF416637 h | MF416442 h |
| Cordyceps takaomontana | BCC12688 | Lepidoptera | EU807996 i | – | – | – | – |
| Cordyceps tenuipes | TBRC7265 | Lepidoptera (pupa) | MF140741 i | MF140707 i | MF140827 i | MF140776 i | MF140800 i |
| TBRC7266 | MF140742 i | MF140708 i | MF140828 i | MF140777 i | MF140801 i | ||
| Engyodontium aranearum | CBS309.85 | Araneae; spider | – | AF339526 c | DQ522341 c | DQ522387 c | DQ522439 c |
| Gibellula pulchra | NHJ10808 | Araneae; spider | – | EU369035 d | EU369018 d | EU369056 d | EU369076 d |
| Gibellula ratticaudata | ARSEF1915 | Araneae; spider | – | DQ518777 d | DQ522360 d | DQ522408 d | DQ522467 d |
| Gibellula sp. | NHJ5401 | Araneae; spider | – | – | – | EU369059 d | EU369097 d |
| Hevansia arachnophila | NHJ10469 | Araneae; spider | – | EU369031 d | EU369008 d | EU369047 d | – |
| Hevansia cinerea | NHJ3510 | Araneae; spider | – | – | EU369009 d | EU369048 d | EU369070 d |
| Hevansia nelumboides | BCC2093 | – | – | MF416530 h | MF416473 h | – | MF416437 h |
| Hevansia novoguineensis | NHJ11923 | Araneae; spider | – | EU369032 d | EU369013 d | EU369052 d | EU369072 d |
| NHJ13161 | Araneae; spider | – | – | EU369011 d | EU369050 d | – | |
| Hevansia websteri | BCC23860 | – | – | – | GQ250030 l | – | – |
| Isaria farinosa | OSC111005 | – | – | DQ518772 h | DQ522348 h | DQ522394 h | – |
| OSC111006 | – | – | EF469080 h | EF469065 h | EF469094 h | – | |
| Isaria sp. | spat09.050 | – | – | MF416559 h | MF416506 h | MF416663 h | MF416457 h |
| spat09.051 | – | – | MF416560 h | MF416507 h | MF416664 h | MF416458 h | |
| Lecanicillium antillanum | CBS350.85 T | Fungi; agaric (Hymenomycetes) | – | AF339536 d | DQ22350 d | DQ522396 d | DQ522450 d |
| Lecanicillium psalliotae | CBS101270 | Soil | – | EF469081 c | EF469066 c | EF469095 c | EF469113 c |
| CBS532.81 | – | AF339560 c | EF469067 c | EF469096 c | EF469112 c | ||
| Purpureocillium lilacinum | CBS284.36 | Soil | AY624189 b | FR775484 c | EF468792 c | EF468898 c | EF468941 c |
| CBS431.87 | Nematoda; Meloidogyne sp. | AY624188 f | EF468844 f | EF468791 f | EF468897 f | EF468940 f | |
| Samsoniella aurantia | TBRC7271 | Lepidoptera | MF140764 i | MF140728 i | MF140846 i | MF140791 i | MF140818 i |
| TBRC7272 | MF140763 i | MF140727 i | MF140845 i | – | MF140817 i | ||
| Samsoniella inthanonensis | TBRC7915 | Lepidoptera (pupa) | MF140761 i | MF140725 i | MF140849 i | MF140790 i | MF140815 i |
| TBRC7916 | MF140760 i | MF140724 i | MF140848 i | MF140789 i | MF140814 i | ||
| Simplicillium lamellicola | CBS116.25 | Soil | AJ292393 f | AF339552 f | DQ522356 f | DQ522404 f | DQ522462 f |
| Simplicillium lanosoniveum | CBS704.86 | Fungi; Hemileia vastatrix | – | AF339553 c | DQ522358 c | DQ522406 c | DQ522464 c |
| CBS101267 | AJ292395 f | AF339554 f | DQ522357 f | DQ522405 f | DQ522463 f | ||
| Simplicillium obclavatum | CBS311.74 | Air above sugarcane field | – | AF339517 c | EF468798 c | – | – |
| Torrubiella wallacei | CBS101237 | Lepidoptera | – | AY184967 c | EF469073 c | EF469102 c | EF469119 c |
| Verticillium sp. | CBS102184 | – | – | AF339564 h | EF468803 h | EF468907 h | EF468948 h |
The phylogenetic analyses were run using a combined dataset comprising four loci: LSU, TEF, RPB1, and RPB2. The combined dataset included 3511 characters, of which 2053 characters were constant, 231 were parsimony-uninformative, and 1227 were parsimony-informative. Gaps were treated as missing data. The maximum parsimony analyses resulted in 31 equally most parsimonious trees, of which one is shown in Figure
Phylogenetic tree based on combined dataset of LSU, TEF, RPB1 and RPB2, sequences showing the relationship of Akanthomyces from Thailand with other species of Cordycipitaceae. Numbers above lines at significant nodes represent Maximum likelihood bootstrap values, Bayesian posterior probabilities, and MP bootstrap values. Bold lines mean support for the three analyses were 100%.
Taxonomy
Akanthomyces noctuidarum , sp. nov.
Type
Thailand. Narathiwat Province, Hala Bala Wildlife Sanctuary, Headquarter Nature Trail; 5°928'N, 101°883'E; on adult moth; 3 Mar 2009; K. Tasanathai (KT), P. Puyngain (PP), T. Chohmee (TC) (holotype BBH 26019 dried culture; ex-type living culture BCC 36265). GenBank: ITS = MT356072, LSU = MT356084, TEF = MT477978, RPB1 = MT477994, RPB2 = MT477987.
Etymology
Referring to the host (Noctuidae, Lepidoptera) where the fungus was found.
Description
Teleomorph: Adult moth attached to the midrib of monocotyledonous leaf or undersides of dicotyledonous leaf covered by white to cream mycelium (OAC816). Stroma arising from host body and wing veins, white to cream, cylindrical, length ca. 5 mm. Perithecia superficial, orange to light brown (OAC825), few to numerous, crowded at the tip of the stroma or growing directly from mycelium in host body and wing veins, ovoid, (530–)623–993(–1000) × (290–)308–413(–425) µm. Asci cylindrical, hyaline, (170–)196–423(–550) × (2–)2.7–3.8(–4) µm. Ascospores cylindrical, filiform, hyaline, multi-septate, breaking into one-celled fragments at maturity, (6–)7–10.7(–13) × 1 µm.
Anamorph: Synnemata arising from moth body and wing veins, white to cream (OAC816), erect, simple, cylindrical to clavate, (650–)668–1191(–1500) × (50–)53.4–102(–120) µm. Conidiogenous cells produced along the synnemata, monophialidic or polyphialidic. Phialides cylindrical with papillate end, hyaline, (5–)6.8–9(–10) × (1.8–) 2–2.4(–3) µm. Conidia cylindrical with round end, hyaline, (3–)3.5–4.5(–6) × 1 µm.
Culture characters
Colony on PDA growing with a diameter of 20–24 mm in 14 days, circular, flat to raised, entire edges, white (OAC909) and fluffy mycelium. Colony reverse cream (OAC814). Colony on OA growing with a diameter of 20–25 mm in 14 days, circular, flat to raised, entire, white (OAC 909) and fluffy mycelium. Colony reverse uncolored. Conidia and reproductive structures not observed on both, PDA and OA in 14 days.
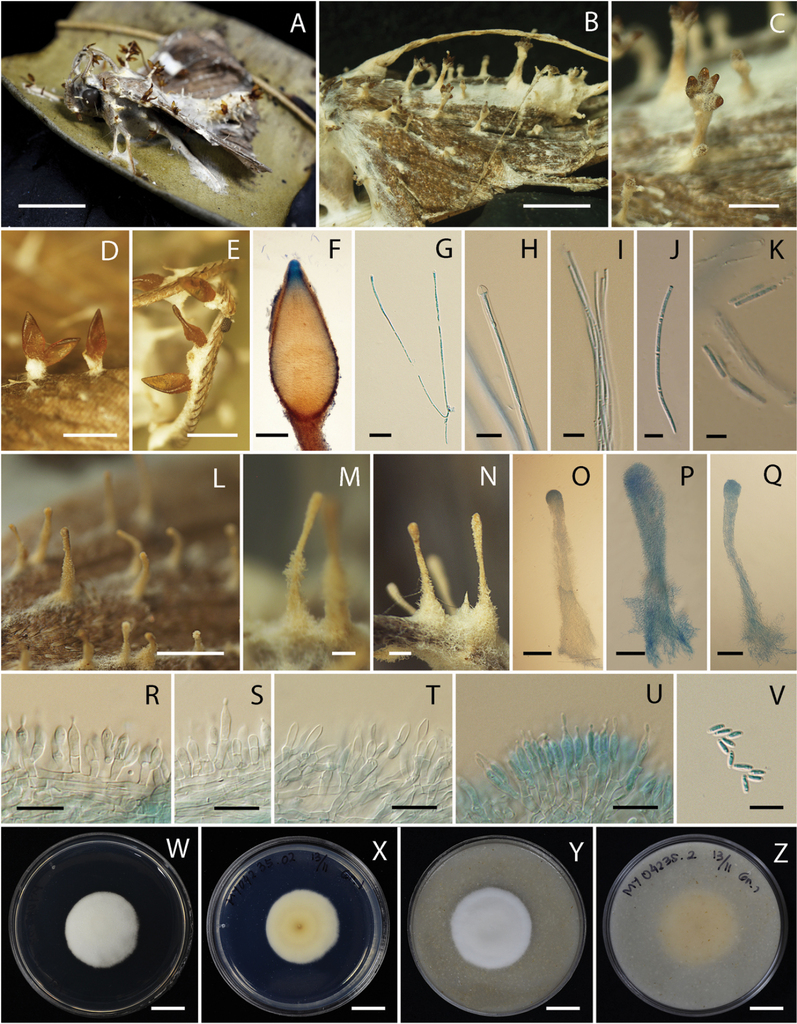
Akanthomyces noctuidarum (BBH 26019, BCC 36265) A, B fungus on adult moth C–F perithecia G asci H tip of ascus I ascus with ascospores J ascospores with clear septae K ascospores break into part-spores L–Q synnemata R–T phialides through the length of synnema U phialides at the tip of synnema V conidia W, X culture on PDA 14 days X reverse Y, Z culture on OA 14 days Z reverse. Scale bars: 1 cm (A, B, W, X, Y, Z); 5 mm (C, I, J, K); 1 mm (D, E, L); 200 µm (F, M, N, O, P, Q); 50 µm (G); 10 µm (H, R, S, T, U, V).
Distribution
Thailand, known from various national parks throughout the country.
Ecology
All specimens were found on the underside of leaves of plants.
Additional specimens examined
Thailand. Nakhon Ratchasima Province, Khao Yai National Park, Km.29; 14°711'N, 101°421'E; on adult moth; 24 Jan 2006; KT, W. Chaygate (WC), S. Sivichai, Le Tan Hung (BBH16595). Narathiwat Province, Hala Bala Wildlife Sanctuary, Headquarter Nature Trail; 5°928'N, 101°883'E; on adult moth; 19 Feb 2011; KT (BBH30267, BCC 47498). Kamphaeng Phet Province, Khlong Lan National Park, Saphan Ton Nature Trail; 16°203'N, 99°321'E; on adult moth; 6 Nov 2007; BT, KT, WC, S. Mongkolsamrit (SM), P. Srikitikulchai (PS), R. Ridkaew (RR), A. Khonsanit (AK) (BBH22738, BCC 28571).
Notes
This species produced both, anamorph and teleomorph. The type strain of this species, BBH 26019/ BCC 36265, consisted of both, anamorph and teleomorph. The other strains produced only one morph on the insect, either anamorph or teleomorph.
Akanthomyces pyralidarum , sp. nov.
Type
Thailand. Kanchanaburi Province, Thung Yai Naresuan Wildlife Sanctuary, Krathon Ruesi Nature Trail; 14°746'N, 98°625'E; on adult moth; 11 Dec 2007; KT, SM, RR, B. Thongnuch (BT) (holotype BBH23823 dried culture, ex-type living culture BCC 28816). GenBank: ITS = MT356080, LSU = MT356091, TEF = MT477982, RPB1 = MT478000, RPB2 = MT478007.
Etymology
Refers to the host (Pyralidae, Lepidoptera) of the fungus.
Description
Teleomorph: Adult moth attached on the undersides of dicotyledonous leaf covered by white to cream mycelium (OAC816). Stroma arising from host body and wings, white to cream (OAC816), cylindrical. Perithecia superficial, crowded at the tip of stroma or growing directly from mycelium that covers the host body, few to numerous, ovoid to obpyriform, (290–)342–580(–650) × (150–)186–291(–340) µm. Asci cylindrical, the bottom of asci thicker than the middle part, (170–)222–329(–360) × (2–)2.5–3.3(–4) µm. Ascospores hyaline, filiform, multi-septate, discharged into part-spores, (5–)5.9–9.4(–12) × 1 µm.
Culture characters
Colonies on PDA growing with a diameter of 23–28 mm in 14 days, white (OAC909), circular, flat, entire. Colony reverse pale yellow (OAC856) at the center. Conidia and reproductive structures not observed. Colonies on OA growing with a diameter of 27–30 mm in 14 days, white (OAC909), circular, flat, entire. Colony reverse uncolored. Conidia and reproductive structures not observed.
Akanthomyces pyralidarum (BBH 23823, BCC 28816) A fungus on adult moth B–F perithecia G, H asci I tip of ascus with immature ascospore J tip of ascus with mature ascospores K ascospores L, M culture on PDA 14 days M reverse N, O culture on OA 14 days O reverse P culture on OA 28 days. Scale bars: 1 cm (A, L, M, N, O, P); 1 mm (B); 500 µm (C, D, E); 100 µm (F); 10 µm (G, H); 5 µm (I, J, K).
Distribution
Thailand, known from various national parks throughout the country.
Ecology
All specimens are found on the underside of leaves of plants.
Additional specimens examined
Thailand. Chiang Mai Province, Huai Nam Dang National Park, Pong Dueat Pa Pae Geyser; 19°121'N, 98°943'E; on adult moth; 5 Sep 2008; KT, WC, PS, AK, SM (BBH 24623, BCC 32191). Phetchabun Province, Nam Nao National Park, Headquarter Nature Trail; 16°768'N, 101°671'E; on adult moth; 24 Nov 2009; KT, TC, AK (BBH 27293, BCC 40869). Kanchanaburi Province, Thung Yai Naresuan Wildlife Sanctuary, Thi Khong Protect Forest Unit; 14°746'N, 98°625'E; on adult moth; 12 Dec 2007; KT, SM, RR, BT (BBH 23778, BCC 29197).
Notes
Akanthomyces pyralidarum is found only in its teleomorph state. This species differs from Akanthomyces noctuidarum by having smaller perithecia (290–650 × 150–340 µm) than A. noctuidarum (530–1000 × 290–425 µm).
Akanthomyces tortricidarum , sp. nov.
Type
Thailand. Nakhon Ratchasima Province, Khao Yai National Park, Mo Sing To Nature Trail; 14°711'N, 101°421'E; on adult moth; 6 Jun 2014; W. Noisripoom, PS, TC, S. Sommai, R. Somnuk (holotype BBH 38669 dried culture, ex-type living culture BCC 72638). GenBank: ITS = MT356076, LSU = MT356088, TEF = MT478004, RPB1 = MT477997, RPB2 = MT477992.
Etymology
Refers to the host (Tortricidae, Lepidoptera) of the fungus.
Description
Anamorph: Specimens examined in this study can be found on the underside of dicotyledonous leaves and palm leaf. The hosts were adult moths, ca. 4–9 × 1–2 mm. Two types of synnemata were produced on insect hosts. Several long synnemata arose at the head and in the middle of the host body, white to cream, up to 5 mm long and ca. 120–150 µm wide, rarely branched, cylindrical to clavate with acute or blunt end. Conidiogenous cells produced along synnemata, monophialidic or polyphialidic. Phialides (5–)6–8(–10) × (1.8–)2–2.7(–3) μm, cylindrical to ellipsoidal with papillate end. Conidia smooth-walled, hyaline, single-celled, fusoid, (2–)2.5–3(–3.2) × (0.8–)1–1.4(–2) µm. Several short synnemata arose on moth body, wings, and legs, white to cream, (197–)200–267(–300) × (15–)17.7–31.6(–40) µm, with diameter of the tip (43–)51.5–73(–75) µm, cylindrical with subglobose or oblong end. Conidiogenous cells produced at the end of synnemata, monophialidic or polyphialidic. Phialides (5–)6.2–8.3(–10) × (1.8–)2–2.5(–3) μm, cylindrical to ellipsoidal with papillate end. Conidia smooth-walled, hyaline, single-celled, fusoid, (1–)1.8–2.7(–3) × 1–2 µm. Phialides and conidia from both long and short synnemata were on the same size range.
Culture characters
Colonies on PDA growing with a diameter of 25–31 mm in 14 days, white (OAC909), circular, flat, entire, reverse pale yellow (OAC858). Mycelium smooth, septate, hyaline. Colonies on OA growing with a diameter of 18–25 mm in 14 days, circular, flat, entire, white (OAC909), reverse brownish yellow (OAC812). Mycelium smooth, septate, hyaline. Conidia and reproductive structures not produced on both, PDA and OA in 14 days.
Akanthomyces tortricidarum (BBH 38669, BCC 72638) A fungus on adult moth B, C, N–P short synnemata D–F long synnemata G–L phialides from long synnema M conidia from long synnema Q–T phialides from short synnema U conidia from short synnema V, W culture on PDA 14 days W reverse X, Y culture on OA 14 days Y reverse. Scale bars: 2 mm (A); 200 µm (B, E); 100 µm (C, F); 500 µm (D); 5 µm (G, M, Q, R, S, T, U); 2 µm (H, I, J, K, L); 30 µm (N, O, P); 1 cm (V, W, X, Y).
Distribution
Thailand, known from various national parks throughout the country.
Ecology
All specimens are found on the underside of leaves of plants.
Additional specimens examined
Thailand. Nakhon Ratchasima Province, Khao Yai National Park, Mo Sing to Nature Trail; 14°711'N, 101°421'E; on adult moth; 7 Apr 2010; KT, SM, TC, AA, RR (BBH 28530, BCC 41868). Nakhon Ratchasima Province, Khao Yai National Park, Mo Sing to Nature Trail; 14°711'N, 101°421'E; on adult moth; 11 Nov 2009; KT, SM, TC, RR, M. Sudhadham, AK (BBH 27283, BCC 40005). Kamphaeng Phet Province, Khlong Lan National Park, Saphan Ton Nature Trail; 16°203'N, 99°321'E; on adult moth; 6 Nov 2007; KT, SM, PS, BT, RR, AK, WC (BBH 23097, BCC 28583).
Notes
Akanthomyces tortricidarum is found only in its anamorph state. This species differs from A. noctuidarum by having smaller conidia (2–3 × 1 µm) than A. noctuidarum (3–6 × 1 µm). Furthermore, the shape of conidia of A. tortricidarum is fusoid, while conidia of A. noctuidarum is cylindrical with a round end.
Discussion
The genus Akanthomyces established by
The type species of Akanthomyces, A. aculeatus and another Akanthomyces species on moth, A. pistillariiformis (= A. tuberculatus), were the closest related species to the three new species described here. Two of three new species were found in their anamorph state. Fortunately, in A. noctuidarum both, teleomorph and anamorph are present in the same specimen. The anamorph comparison between some species within Akanthomyces is shown in Table 2. The conidia of A. noctuidarum and A. aculeatus are almost in the same size (A. noctuidarum; 3–6 × 1 µm, A. aculeatus; 3–6 × 2–3 µm). However, the conidial shape of A. noctuidarum is cylindrical with a round end while A. aculeatus is ellipsoid or obovoid. Akanthomyces noctuidarum has the smallest synnemata compared to all the others (A. noctuidarum; 650–1500 µm, A. aculeatus; 1–8 × 0.1–0.5 mm, A. tuberculatus; 1–6 mm × 50–300 µm). Akanthomyces noctuidarum also has smaller phialides than both aforementioned species (5–10 × 2–3 µm, A. aculeatus; 6–16 × 2.5–4 µm, A. tuberculatus; 7–10.5 × 2.7–3.5 µm) with cylindrical shape and papillate at the end.
Morphological comparisons between anamorph of closely related Akanthomyces species used in this study.
| Species | Host | Synnemata | Phialides | Conidia |
|---|---|---|---|---|
| Akanthomyces aculeatus 2 | Moth (Lepidoptera) | Yellowish, cylindrical, narrowing upward, 1–8 mm long and 0.1–0.5 mm wide | Subcylindric to narrowly ellipsoidal, 6–16 × 2.5–4 µm | Ellipsoidal or obovoid, 3–6 × 2–3 µm |
| Akanthomyces angustispora 2 | Coleoptera larva | Flesh colored, simple or branched, 8–13 mm long and 0.2–0.6 mm wide | Oblong or narrowly ellipsoidal, 6–14 × 3–4 µm | Narrowly clavate, 4.5–6 × 1.2–1.4 µm |
| Akanthomyces arachnophilus 4 | Spider (Araneae) | Creamish yellow to pale brown, simple or branched, cylindrical, 2.5–5 mm × 50–75 µm | Globose, 3.2–4.3 × 6.5–8.5 µm | Fusiform, 4.5–5.5(–6) × 1.5–3 µm |
| Akanthomyces araneogenum 5 | Spider | Conidiophores mononematous or synnematous, 21.6–48 × 1.2–2.2 μm, penicillium-like from hyphae directly | Cylindrical, somewhat inflated base, tapering to a thin neck, 4.3–17.3 × 0.9–3.1 μm | Globose, 1.3–2.4 μm in diam, or ellipsoidal, 2.1–3.3 × 1.1–1.6 μm |
| Akanthomyces gracilis 4 | Hymenoptera, Coleoptera, Lepidoptera (moth larvae) Heteroptera, Homoptera | White to yellow-brown, simple, rarely branched, cylindrical, usually 0.7–2 mm × 100–400 µm, occasionally up to 30 mm long and 0.5 mm wide | Cylindrical, 7–10 × 1.5–2.5 µm | Ellipsoidal, fusiform, 2.5–3 × 1–1.6 µm |
| Akanthomyces kanyawimiae 3 | Spider (Araneae) | Up to 1.5 mm long, up to 400 µm wide; loosely covered by dense white to cream mycelia | Cylindrical to ellipsoidal, (7–)8–10.5(–12) × 2–3 µm | Fusiform or lemon-shaped, (2–)2.5–3.5(–4) × 1–2 µm |
| Akanthomyces noctuidarum 1 | Lepidoptera; Noctuidae | White to cream (OAC816), simple, cylindrical to clavate, (650–)668–1191(–1500) × (50–)53–102(–120) µm | Cylindrical with papillate end, (5–)6.8–9(–10) × (1.8–)2–2.4(–3) µm | Cylindrical with round end, (3–)3.5–4.7(–6) × 1 µm |
| Akanthomyces pistillariiformis 4 (= A. tuberculatus) | Moth (Lepidoptera) | White to creamish, simple, occasionally branched, cylindrical to clavate and stipitate, 1–6 mm long and 50–300 µm wide | Cylindrical, 7–10.5 × 2.7–3.5 µm | Cylindrical to narrowly fusiform, 4.5–6 × 1.2–1.5 µm |
| Akanthomyces suphureus 3 | Spider (Araneae) | – | Cylindrical, (5–)7.5–11(–12) × 2–2.5 µm | Cylindrical to ellipsoidal, (3–)4(–5) × (1–)1.5(–2) µm |
| Akanthomyces tortricidarum 1 | Lepidoptera; Tortricidae | Long synnemata white to cream, rarely branched, cylindrical to clavate with acute or blunt end, up to 5 mm long and wide ca. 120–150 µm. | Cylindrical to ellipsoidal with papillate end, (5–)6–8(–10) × (1.8–)2–2.7(–3) μm | Fusoid, (2–)2.5–3(–3.2) × 1–2 µm |
| Short synnemata white to cream, cylindrical with subglobose or oblong at the end, (197–)200–267(–300) × (15–)17.7–31.6(–40) µm, with diameter of the tip (43–)51.5–73(–75) µm | Cylindrical to ellipsoidal with papillate end, (5–)6.2–8.3(–10) × (1.8–)2–2.5(–3) μm | Fusoid, (1–)1.8–2.7(–3) × 1–2 µm | ||
| Akanthomyces waltergamsii 3 | Spider (Araneae) | White to cream synnemata up to 1.5 mm long and ca. 100–120 µm wide | Cylindrical to ellipsoidal, (7–)8.5–11(–12) × 2.5–3 µm | Ellipsoidal, fusiform, (3–)4–5.5(–6) × 1.5–2 µm |
Akanthomyces tortricidarum was distinguished from the others species by having two different types of synnemata. The long synnemata of A. tortricidarum are cylindrical to clavate with acute or blunt ends. The hyphae diverged in the upper portion of the synnema and repeatedly branched more or less dichotomously, whereas the phialides were terminal on the branches. At the lower portion of synnema, the phialides were produced either as lateral cells or frequently as terminal cells of short lateral branches produced along the entire length of the outer hyphae of the synnema. The production of phialides was abundant at the upper portion of the synnema, resulting in a compact hymenial layer, whereas the phialides at the lower portion of the synnema were scattered and well separated from each other. Unlike the long synnemata, the hymenium-like layer of phialides on the short synnemata was limited to its upper part and the lower portion was sterile, forming a stipe. In the upper portion of the short synnema, the hyphae diverged and repeatedly branched more or less dichotomously and terminated with phialides. However, at the lower portion, the outer longitudinal hyphae did not produce any lateral phialides or lateral branches bearing phialides. This character was similar to the genus Insecticola proposed by
The teleomorph comparison between some species within Akanthomyces is shown in Table 3. Akanthomyces noctuidarum and A. pyralidarum differed from A. tuberculatus by the size of ascospores, asci, and perithecia. Akanthomyces tuberculatus has smaller ascospores measuring 2–6 × 0.5–1 µm, whereas A. noctuidarum and A. pyralidarum have larger ascospores at 6–13 × 1 µm and 5–12 × 1 µm, respectively. However, all three of them have the same shape of ascospore and asci. Akanthomyces pyralidarum has the smallest size of asci (A. pyralidarum; 170–360 × 2–4 µm, A. noctuidarum; 170–550 × 2–4 µm, and A. tuberculatus; 300–600 × 4–5 µm). The shape of perithecia from A. noctuidarum is ovoid, while A. pyralidarum is ovoid to obpyriform and A. tuberculata is narrowly ovoid or conoid. Akanthomyes pyralidarum also has the smallest perithecia compared to the other species in the genus (A. pyralidarum; 290–650 × 150–340 µm, A. tuberculatus; 420–900 × 180–370 µm, A. sulphureus; 650–680 × 240–330 µm, A. thailandicus; 700–850 × 300–400 µm, and A. noctuidarum; 530–1000 × 290–425 µm). Moreover, A. sulphureus and A. thailandicus are found on spiders (Araneae) while the others were found on moths.
All strains from these species did not produce conidia or reproductive structures when grown on PDA and OA for 14 days at 25 °C. Nevertheless, one strain from A. pyralidarum (BCC 29197) started to produce a synnemata-like structure on OA after 28 days. However, this synnemata-like structure was sterile and did not produce any phialides or conidia. Overall, fungal growth was faster in OA than in PDA.
Morphological comparisons between teleomorph of closely related Akanthomyces species used in this study.
| Species | Host | Perithecia | Asci | Ascospores |
|---|---|---|---|---|
| Akanthomyces noctuidarum 1 | Lepidoptera; Noctuidae | Superficial, orange to light brown, ovoid, (530–)623–993(–1000) × (290–)308–413(–425) µm | Cylindrical, (170–)196–423(–550) × (2–)2.7–3.8(–4) µm | Cylindrical, filiform, multi-septate, part-spores, (6–)7–10.7(–13) × 1 µm |
| Akanthomyces pyralidarum 1 | Lepidoptera; Pyralidae | Superficial, ovoid to obpyriform, (290–)342–580(–650) × (150–)186–291(–340) µm | Cylindrical, (170–)222–329(–360) × (2–)2.5–3.3(–4) µm | Filiform, multi-septate, part-spores, (5–)5.9–9.4(–12) × 1 µm |
| Akanthomyces suphureus 2 | Spider (Araneae) | Superficial, ovoid, (650–)676(–680) × (240–)324.5(–330) µm | Cylindrical, up to 500 µm long, 2–3 µm wide | Whole, filiform, (300–) 336(–450) × 1–1.5 µm |
| Akanthomyces thailandicus 2 | Spider (Araneae) | Superficial, narrowly ovoid, (700–)752–838(–850) × (300–)305–375(–400) µm | Cylindrical, up to 550 µm long, 5–7 µm wide | Cylindrical, multi-septate, part-spores, 4–6 × 1–1.5 µm |
| Akanthomyces tuberculatus3 (= C. tuberculata) | Moth (Lepidoptera) | Superficial, narrowly ovoid or conoid, dark brown, 420–900 × 180–370 µm | Cylindrical, 300–600 × 4–5 µm with a 4 µm thick cap | Filiform, multi-septate, part-spores, 2–6 × 0.5–1 µm |
Acknowledgements
This research was supported by the Platform Technology Management Section, National Center for Genetic Engineering and Biotechnology (BIOTEC), Grant No. P19-50231, National Science and Technology Development Agency (NSTDA), Thailand. We thank Natnapha Phosrithong for insect identification. We are indebted to the Department of National Parks, Wildlife and Plant Conservation of Thailand for their cooperation and support for our project research. Arifah Nur Aini and Anto Budiharjo thank to Diponegoro University - Indonesia for providing RPIBT grant No. 387–03/UN7.P4.3/PP/2018.
References
- Afandhi A, Widjayanti T, Emi AAL, Tarno H, Afiyanti M, Handoko RNS (2019) Endophytic fungi Beauveria bassiana balsam accelerates growth of common bean (Phaeseolus vulgaris L.). Chemical and Biological Technologies in Agriculture 11(6): 2–6. https://doi.org/10.1186/s40538-019-0148-1
- Castlebury LA, Rossman AY, Sung GH, Hyten AS, Spatafora JW (2004) Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycological Research 108(8): 864–872. https://doi.org/10.1017/S0953756204000607
- Chaverri P, Bischoff JF, Evans HC, Hodge KT (2005) Regiocrella, a new entomopathogenic genus with a pycnidial anamorph and its phylogenetic placement in the Clavicipitaceae. Mycologia 97(6): 1225–1237. https://doi.org/10.1080/15572536.2006.11832732
- Chen W-H, Liu C, Han Y-F, Liang J-D, Liang Z-Q (2018) Akanthomyces araneogenum, a new isaria-like araneogenous species. Phytotaxa 379(1): 66–72. https://doi.org/10.11646/phytotaxa.379.1.6
- Chiriví-Salomón JS, Danies G, Restrepo S, Sanjuan T (2015) Lecanicillium sabanense sp. nov. (Cordycipitaceae) a new fungal entomopathogen of coccids. Phytotaxa 234(1): 63–74. https://doi.org/10.11646/phytotaxa.234.1.4
- Das SK, Masuda M, Sakurai A, Sakakibara M (2010) Medicinal uses of the mushroom Cordyceps militaris: current state and prospects. Fitoterapia 81(8): 961–968. https://doi.org/10.1016/j.fitote.2010.07.010
- Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5): 1792–1797. https://doi.org/10.1093/nar/gkh340
- Gams W, Zare R (2001) A revision of Verticillium sect. Prostrata. III. Generic classification. Nova Hedwigia 72: 329–337.
- Hall T (2005) BioEdit, version 7.0.5.3. Raleigh, North Carolina: Department of Microbiology, North Carolina State University. http://www.mbio.ncsu.edu/bioedit.html
- Johnson D, Sung GH, Hywel-Jones NL, Luangsa-ard JJ, Bischoff JF, Kepler RM, Spatafora JW (2009) Systematics and evolution of the genus Torrubiella (Hypocreales, Ascomycota). Mycological Research 113: 279–289. https://doi.org/10.1016/j.mycres.2008.09.008
- Kepler RM, Sung G-H, Ban S, Nakagiri A, Chen MJ, Huang B, Li Z, Spatafora JW (2012) New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia 104(1): 182–197. https://doi.org/10.3852/11-070
- Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, Quandt CA, Sung GH, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, Chen M, Li Z, Rossman AY, Spatafora JW, Shrestha B (2017) A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8(2): 335–353. https://doi.org/10.5598/imafungus.2017.08.02.08
- Kim YO, Kim HJ, Abu-taweel GM, Oh J, Sung GH (2018) Neuroprotective and therapeutic effect of Cordyceps militaris on ischemia-induced neuronal death and cognitive imapirments. Saudi Journal of Biological Sciences 26: 1352–1357. https://doi.org/10.1016/j.sjbs.2018.08.011
- Kobmoo N, Mongkolsamrit S, Tasanathai K, Thanakitpipattana D, Luangsa-ard JJ (2012) Molecular phylogenies reveal host-specific divergence of Ophiocordyceps unilateralis sensu lato following its host ants. Molecular Ecology 21(12): 3022–3031. https://doi.org/10.1111/j.1365-294X.2012.05574.x
- Kuephadungphan W, Macabeo APG, Luangsa-ard JJ, Tasanathai K, Thanakitpipattana D, Phongpaichit S, Yuyama K, Stadler M (2018) Studies on the biologically active secondary metabolites of the new spider parasitic fungus Gibellula gamsii. Mycological Progress 18: 135–146. https://doi.org/10.1007/s11557-018-1431-4
- Læssøe T, Srikitikulchai P, Luangsa-ard JJ, Stadler M (2013) Theissenia reconsidered, including molecular phylogeny of the type species T. pyrenocrata and a new genus Durotheca (Xylariaceae, Ascomycota). IMA Fungus 4(1): 57–69. https://doi.org/10.5598/imafungus.2013.04.01.07
- Lebert H (1858) Über einige neue oder unvollkommen gekannte Krankheiten der Insekten, welche durch Entwicklung niederer Pflanzen im lebenden Körper enstehen. Zeitschrift für wissenschaftliche Zoologie 9: 439–453.
- Liu YJ, Whelen S, Hall BD (1999) Phylogenetic relationships among Ascomycetes: evidence from an RNA Polymerase II subunit. Molecular Biology and Evolution 16(12): 1799–1808. https://doi.org/10.1093/oxfordjournals.molbev.a026092
- Luangsa-ard JJ, Hywel-Jones NL, Samson RA (2004) The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia 96(4): 773–780. https://doi.org/10.1080/15572536.2005.11832925
- Luangsa-ard JJ, Hywel-Jones NL, Manoch L, Samson RA (2005) On the relationships of Paecilomyces sect. Isarioidea species. Mycological Research 109(5): 581–589. https://doi.org/10.1017/S0953756205002741
- Luangsa-ard JJ, Tasanathai K, Thanakitpipattana D, Khonsanit A, Stadler M (2018) Novel and interesting Ophiocordyceps spp. (Ophiocordycipitaceae, Hypocreales) with superficial perithecia from Thailand. Studies in Mycology 89: 125–142. https://doi.org/10.1016/j.simyco.2018.02.001
- Mains EB (1950) Entomogenous species of Akanthomyces, Hymenostilbe, and Insecticola in North America. Mycologia 42(4): 566–589. https://doi.org/10.1080/00275514.1950.12017861
- Mantzoukas S, Lagogiannis I (2019) Endophytic colonization of pepper (Capsicum annum) controls aphids (Myzus persicae Sulzer). Applied Science 9: 2–12. https://doi.org/10.3390/app9112239
- Mongkolsamrit S, Noisripoom W, Thanakitpipattana D, Wuthikun T, Spatafora JW, Luangsa-ard JJ (2018) Disentangling cryptic species with isaria-like morphs in Cordycipitaceae. Mycologia 110(1): 230–257. https://doi.org/10.1080/00275514.2018.1446651
- Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- Rehner B, Buckley E (2005) A Beauveria phylogeny inferred from ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97(1): 84–98. https://doi.org/10.3852/mycologia.97.1.84
- Rehner SA, Minnis AM, Sung GH, Luangsa-Ard JJ, Devotto L, Humber RA (2011) Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103(5): 1055–1073. https://doi.org/10.3852/10-302
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
- Samson RA, Evans HC (1974) Notes on entomogenous fungi from Ghana II. The genus Akanthomyces. Acta Botanica Neerlandica 23(1): 28–35. https://doi.org/10.1111/j.1438-8677.1974.tb00913.x
- Sanjuan T, Tabima J, Silvia R, Læssøe T, Spatafora JW, Franco-Molano AE (2014) Entomopathogens of Amazonian stick insects and locusts are members of the Beauveria species complex (Cordyceps sensu stricto). Mycologia 106(2): 260–275. https://doi.org/10.3852/13-020
- Shrestha B, Kubatova A, Tanaka E, Oh J, Yoon DH, Sung JM, Sung GH (2019) Spider-pathogenic fungi within Hypocreales (Ascomycota): their current nomenclature, diversity, and distribution. Mycological Progress 18: 983–1003. https://doi.org/10.1007/s11557-019-01512-3
- Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
- Sukarno N, Kurihara Y, Ilyas M, Mangunwardoyo W, Yuniarti E (2009) Lecanicillium and Verticillium species from Indonesia and Japan including three new species. Mycoscience 50: 369–379. https://doi.org/10.1007/S10267-009-0493-1
- Sung GH, Spatafora JW, Zare R, Hodge KT, Gams W (2001) A revision of Verticillium sect. Prostrata. II. Phylogenetic analyses of SSU and LSU nuclear rDNA sequences from anamorphs and teleomorphs of the Clavicipitaceae. Nova Hedwigia 72(3): 311–328.
- Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57: 5–59. https://doi.org/10.3114/sim.2007.57.01
- Swofford DL (2019) PAUP: phylogenetic analysis using parsimony. Version 4.0a116. Sunderland, Massachusetts: Sinauer Associates.
- Takakura K, Ito S, Sonoda J, Tabata K, Shiozaki M, Nagai K, Shibata M, Koike M, Uchiyama Y, Gotow T (2017) Cordyceps militaris improves the survival of Dahl salt-sensitive hypertensive rats possibly via influences of mitochondria and autophagy functions. Heliyon 3: e00462. https://doi.org/10.1016/j.heliyon.2017.e00462
- Tasanathai K, Noisripoom W, Chaitika T, Khonsanit A, Hasin S, Luangsa-ard JJ (2019) Phylogenetic and morphological classification of Ophiocordyceps species on termites from Thailand. MycoKeys 56: 101–129. https://doi.org/10.3897/mycokeys.56.37636
- Vega FE, Meyling NV, Luangsa-ard JJ, Blackwell M (2012) Fungal entomopathogens. In: Vega FE, Kaya HK (Eds) Insect Pathology 171–220. https://doi.org/10.1016/B978-0-12-384984-7.00006-3
- Vilgalys R, Sun BL (1994) Ancient and recent patterns of geographic speciation in the oyster mushroom Pleurotus revealed by phylogenetic analysis of ribosomal DNA sequences. Proceedings of the National Academy of Sciences of the United States of America 91: 4599–4603. https://doi.org/10.1073/pnas.91.10.4599
- Vu D, Groenewald M, de Vries M, Gehrmann T, Stielow B, Eberhardt U, Al-Hatmi A, Groenewald JZ, Cardinali G, Houbraken J, Boekhout T, Crous PW, Robert V, Verkley GJM (2019) Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology 92: 135–154. https://doi.org/10.1016/j.simyco.2018.05.001
- White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Snisky JJ, White TJ (Eds) PCR Protocols: a guide to methods and applications. Academic Press, San Diego, California, 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Supplementary materials
MP tree
Data type: phylogenetic tree
Explanation note: Branches showing Maximum Parsimony bootstrap values.
RAxML tree
Data type: phylogenetic tree
Explanation note: Branches showing Maximum Likelihood bootstrap values from RAxML.