A molecular phylogeny of Graphidaceae (Ascomycota, Lecanoromycetes, Ostropales) including 428 species
1 Department of Botany, The Field Museum, 1400 South Lake Shore Drive, Chicago, Illinois 60605-2496, U.S.A.
2 Department of Biology, Duke University, 125 Science Drive, Durham, North Carolina 27708, U.S.A.
3 University of Regensburg, Institute for Botany, Universitätsstr. 31, D-93040 Regensburg, Germany
4 Universität Duisburg-Essen, Botanisches Institut, Universitätsstr. 5, D-45141 Essen, Germany
5 National Museum of Nature and Science, 4-1-1 Amakubo, Tsukuba, Ibaraki, 305-0005 Japan
6 Department of Botany, University of Sri Jayawardenepura, Sri Lanka
7 Fundación Instituto Botánico de Venezuela, Ave. Salvador Allende, Jardín Botánico de Caracas, Universidad Central de Venezuela, Apartado # 2156, Caracas 1010-A, Venezuela
8 Departamento de Biociências, Universidade Federal de Sergipe, CEP: 49.500-000, Itabaiana, Sergipe, Brasil
9 Lichenologisches Institut Neumarkt, Im Tal 12, D-92318 Neumarkt, Germany
10 Botanischer Garten and Botanisches Museum Berlin Dahlem, Königin-Luise-Strasse 6–8, D-14195 Berlin, Germany
11 534 Fenton St., Lansing, Michigan 48910, U.S.A.
12 Committee on Evolutionary Biology, University of Chicago, 1025 E. 57th Street, Chicago, Illinois 60637, U.S.A
Introduction
Graphidaceae, which is currently accepted to include the previously independent families Thelotremataceae (Mangold et al. 2008a, Rivas Plata et al. 2012a), Gomphillaceae, Asterothyriaceae, and Solorinellaceae (Baloch et al. 2010, Rivas Plata et al. 2012a, b), is the second largest family of lichenized fungi and the dominant element of lichen communities in tropical regions, with over 1800 accepted species (Staiger 2002, Frisch et al. 2006a, Archer 2006, 2007, 2009, Lücking and Rivas Plata 2008, Lücking 2009, Lücking et al. 2008, 2009, Rivas Plata et al. 2008, 2012a, Mangold et al. 2009). Quantitative extrapolations and molecular data available for species complexes, as well as continuous new discoveries, suggest that the family may actually contain well over 2000 species (Lücking 2012, Rivas Plata and Lücking 2012, Rivas Plata et al. 2012b, Sipman et al. 2012).
For over 100 years, family and genus concepts in this group were based on apothecia and ascospore types, separating most of the species into groups with either rounded, lirellate, or pseudostromatic ascomata and, for each ascoma type, into four genera depending on whether ascospores were transversely septate or muriform and hyaline or pigmented (Müller 1887, Hale 1974, 1978, Wirth and Hale 1963, 1978, Staiger 2002, Frisch et al. 2006a): rounded (Thelotremataceae): Ocellularia, Thelotrema, Phaeotrema, Leptotrema; Graphidaceae: lirellate (Graphidaceae): Graphis, Graphina, Phaeographis, Phaeographina; pseudostromatic (Graphidaceae): Glyphis, Medusulina, Sarcographa, Sarcographina). This concept was highly schematic and challenged by several authors, e.g. in the former Thelotremataceae by Salisbury (1971, 1972, 1978) and Hale (1980, 1981). The latter author proposed an alternative generic concept with three genera based on excipular structures: carbonized and lacking periphysoids (Ocellularia), non-carbonized and lacking periphysoids (Myriotrema), and non-carbonized and with periphysoids (Thelotrema). However, delimitation of very large genera with well over 300 species each and the problems with generic assignment of aberrant taxa made this concept unsatisfactory. In addition, subsequent phylogenetic studies showed that these genera were not monophyletic (Frisch et al. 2006a, Mangold et al. 2008a). Also, no comparable solution was proposed for lirellate and stromatic species classified in Graphidaceae, which until recently remained essentially unchanged from ascospore-based concepts established in the 19th century (Wirth and Hale 1963, 1978, Archer 1999, 2000, 2001a-d, 2002, Staiger 2002).
Major systematic revisions of both families only started after turn of the millenium, with two monographic treatments put forward by Staiger (2002) and Frisch et al. (2006a). In these, a more detailed approach to genus classification was employed, based on a combination of characters such as thallus structure, excipular structure, hamathecium type, ascospore type, and secondary chemistry. This refined classification, which resulted in many new genera (Staiger 2002; Frisch et al. 2006a, Kalb 2009, Rivas Plata et al. 2010a, Lücking et al. 2011, Cáceres et al. 2012, Sipman et al. 2012) has subsequently been tested using molecular approaches, with a growing amount of sequence data available (Staiger 2002, Kalb et al. 2004, Lumbsch et al. 2008, Frisch et al. 2006b, Staiger et al. 2006, Mangold et al. 2008a, Baloch et al. 2010, Rivas Plata and Lumbsch 2011, Rivas Plata et al. 2012a-b). Using molecular data, it was also shown that Graphidaceae and Thelotremataceae were non-monophyletic and consequently Thelotremataceae was included in Graphidaceae. Within Graphidaceae five major clades were distinguished based on supported monophyletic clades (Mangold et al. 2008a, Rivas Plata and Lumbsch 2011, Rivas Plata et al. 2012a). In addition, two separate studies supported three further families, Gomphillaceae, Asterothyriaceae, and Solorinellaceae, as part of Graphidaceae (Baloch et al. 2010, Rivas Plata et al. 2012a, b). The inclusion of the latter has been controversely discussed (Aptroot 2012, Rivas Plata et al. 2012a), since it appears to disrupt the more or less morphologically homogeneous entity formed by subfamilies Fissurinoideae and Graphidoideae; on the other hand, as pointed out by Sipman et al. (2012), the differences between Gomphilloideae and the remaining Graphidaceae are merely gradual in nature. It can be argued that Graphidaceae could be retained as a paraphyletic entity excluding Gomphillaceae (Aptroot 2012). However, the known topology does not support this view since Gomphilloideae is strongly supported as sister to Fissurinoideae on a long branch, i.e. Fissurinoideae is more closely related to Gomphilloideae than to Graphidoideae and hence, Graphidaceae after exclusion of Gomphillaceae would be polyphyletic rather than paraphyletic. In the present study, subfamily Gomphilloideae is not treated since only few species have been sequenced and this clade will be dealt with in a separate contribution.
In this paper, we present the most recent molecular phylogeny of Graphidaceae, focusing on subfamilies Fissurinoideae and Graphidaceae, using a supermatrix approach of all available and newly generated sequences of the mtSSU, nuLSU rDNA, and RPB2 gene partitions. Based on the results, we discuss the progress and problems in the delimitation of infra-family and genus-level taxa in the family. We did not include subfamily Gomphilloideae in this analysis since this subfamily will be treated in detail in a separate paper and its inclusion or exclusion does not affect the topology of subfamilies Fissurinoideae and Graphidoideae.
Material and methods
Partial sequences of the mtSSU rDNA, nuLSU rDNA and the RPB2 gene were obtained from 907 ingroup OTUs representing 428 species in Graphidaceae (subfamilies Fissurinoideae and Graphidoideae). Thirteen additional species from the ostropalean families Stictidaceae, Porinaceae, Gyalectaceae, and Coenogoniaceae, were used as outgroup. A total of 865 new ingroup sequences were newly generated (Appendix 1): 481 for the mtSSU, 260 for the nuLSU, and 124 for the RPB2 gene. In addition, 433 ingroup sequences were downloaded from GenBank (Appendix 1): 263 for the mtSSU, 151 for the nuLSU, and 19 for the RPB2 gene. Unfortunately, for many OTUs, sequence representation was incomplete, either because original DNA extracts from GB sequences were unavailable or because PCR did not work, especially with nuLSU and RPB2 primers. Thus, of a potential total of 907 × 3 = 2721 ingroup sequences, 1298 (48%) were assembled in the dataset. The mtSSU was most complete (744 out of 907 or 82%), followed by the nuLSU (411 out of 907 or 45%) and RPB2 (143 out of 907 or 16%). A total of 35 further sequences, particularly of the nuLSU, could not be used because of contaminants or else unexplained conflict.
New sequences were generated for this study using the Sigma REDExtract-N-Amp Plant PCR Kit (St. Louis, Missouri, SA) for DNA isolation following the manufacturer’s instructions, except that 40 μL of extraction buffer and 40 μL dilution buffer were used. DNA dilutions (5x) were used in PCR reactions of the genes coding for the nuLSU, mtSSU and RPB2, respectively. Primers for amplification were: (a) for nuLSU: AL2R (Mangold et al. 2008a), and nu-LSU-1125-3’ (= LR6) (Vilgalys and Hester 1990), (b) for mtSSU: mr-SSU1 and Mr-SSU3R (Zoller et al. 1999), and (c) for RPB2: fRPB2-7cF and fRPB2-11aR (Liu et al. 1999). PCR reactions contained 5.0 μL R4775 Sigma REDExtract-N-Amp™ PCR ReadyMix, 0.5 μL of each primer (10 μM), 2 μL genomic DNA extract and 2 μL distilled water for a total of 10 μL. Thermal cycling parameters were: (1) for nuLSU: initial denaturation for 5 min at 94°C, followed by 35 cycles of 30 s at 95°C, 30 s at 58°C, 1 min at 72°C, and a final elongation for 10 min at 72°C; (2) for mtSSU: initial denaturation for 5 min at 95°C, followed by 35 cycles of 45 s at 94°C, 1 min at 50°C, 1 min 30 s at 72°C, and a final elongation for 10 min at 72°C; and (3) for RPB2: initial denaturation for 3 min at 95°C, then 1 min at 95°C, and 37 cycles of 1 min at 57°C, 1 min at 58°C, 1 min at 59°C, 1 min at 60°C, 1 min at 61°C, 1 min at 62°C, 1 min at 63°C, 1 min at 64°C and 1.5 min at 72°C, and a final elongation for 10 min at 72°C. Samples were visualized on a 1% ethidium bromide-stained agarose gel under UV light and bands were gel extracted, heated at 70° C for 5 minutes, cooled to 45° C for 10 minutes, treated with 1 μL GELase (Epicentre Biotechnologies, Madison, WI, USA) and incubated at 45° C for at least 24 hours. The 10 μl cycle sequencing reactions consisted of 1–1.5 μl of Big Dye version 3.1 (Applied Biosystems, Foster City, California, U.S.A.), 2.5–3 μl of Big Dye buffer, 6 μM primer, 0.75–2 μl gelased PCR product and water. Samples were sequenced with PCR primers. The cycle sequencing conditions were as follows: 96° C for 1 minute, followed by 25 cycles of 96° C for 10 seconds, 50° C for 5 seconds and 60° C for 4 minutes. Samples were precipitated and sequenced using Applied Biosystems 3730 DNA Analyzer (Foster City, California, U.S.A.), sequences were assembled in SeqMan 4.03 (DNASTAR) and submitted to GenBank (Appendix 1).
Sequences were arranged into multiple sequence alignments (MSA) for each gene using BioEdit 7.09 (Hall 1999) and automatically aligned with MAFFT using the --auto option (Katoh and Toh 2005). The unaligned MSA for the mtSSU and nuLSU gene partitions were also submitted to the GUIDANCE web server at http://guidance.tau.ac.il to assess alignment confidence scores for each site (Penn et al. 2010a, b). GUIDANCE uses a MAFFT alignment and returns a colored MSA that allows delimiting ambiguously aligned portions of the MSA. These were then excluded from further analysis. Introns were deleted from the nuLSU gene partition because of their random occurrence but kept in the mtSSU partition if consistent within species or species groups. This resulted in alignments of 995 sites for the mtSSU, 972 sites for the nuLSU, and 951 for RPB2, for a total of 2918 sites in the combined dataset. After testing for supported topological conflicts (Mason-Gamer and Kellogg 1996, Miadlikowska and Lutzoni 2000, Kauff and Lutzoni 2002), the three genes were combined into a single supermatrix. Individual datasets and the combined supermatrix were subjected to maximum likelihood search using the RAxML-HPC BlackBox 7.3.2 on the Cipres Gateway server (Stamatakis 2006, Stamatakis et al. 2005, 2008; Miller et al. 2010; http://www.phylo.org/portal2/login !input.action), with parametric bootstrapping generating 350 replicates as automatically determined by RAxML using a saturation criterion. The universal GTR-Gamma model was chosen for the analysis.
Results
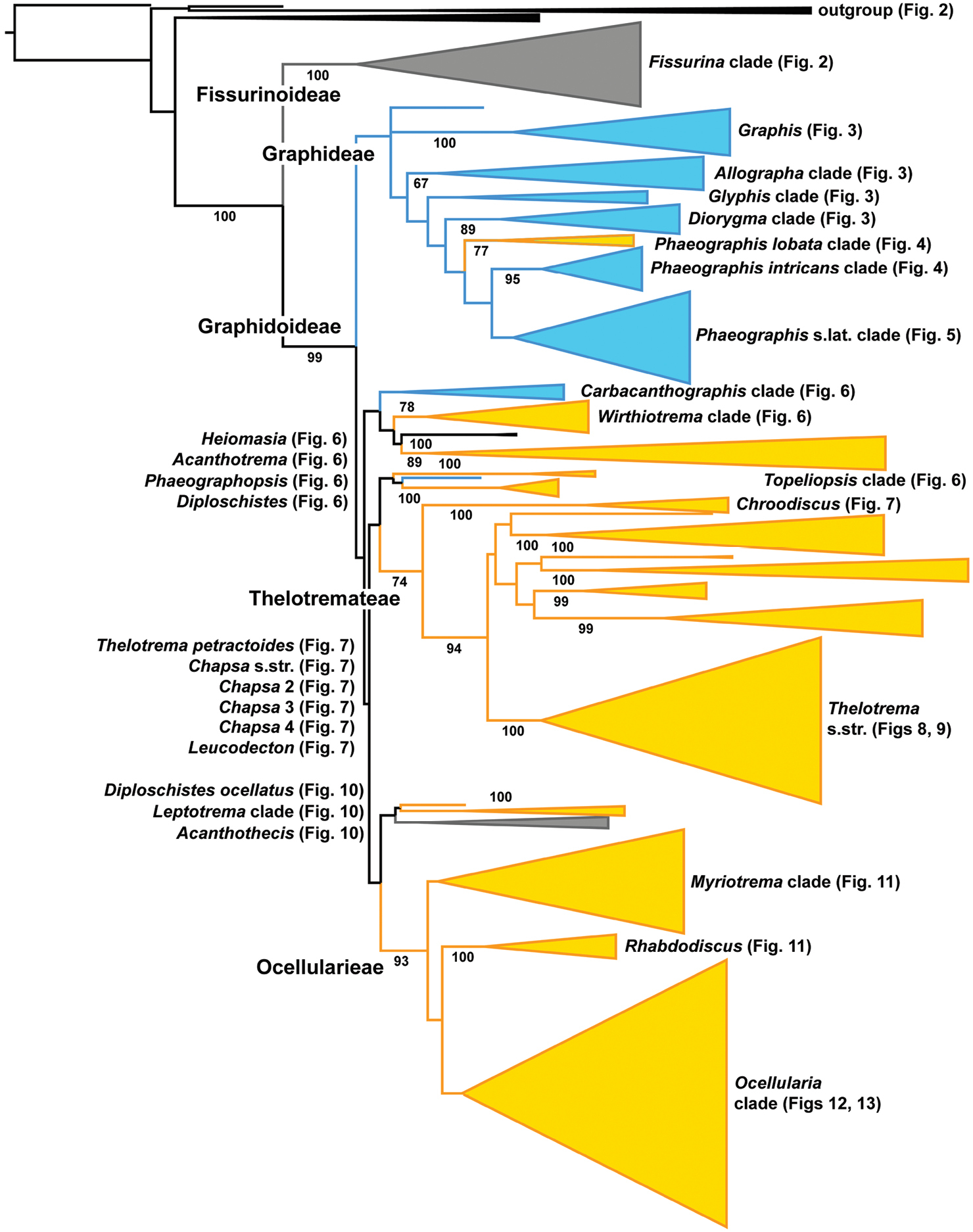
In our analysis, the core Graphidaceae is divided into two strongly supported clades representing subfamilies Fissurinoideae and Graphidoideae (Fig. 1, see Appendix 2 for entire, fully resolved tree). Subfamily Graphidoideae is further divided into six larger and smaller clades, some of which form unsupported clusters. These clades largely represent species with either lirellate or rounded ascomata, but no supported division into two clades representing either ascomata type is evident; also, subfamily Fissurinoideae includes both species with lirellate and rounded ascomata.
Figure 1.
Cartoon tree showing the major clades distinguished within Graphidaceae with bootstrap support given next to branches. Blue clades indicate graphidoid taxa (lirellate or pseudostromatic ascomata), orange clades indicate thelotremoid taxa (rounded ascomata), and grey clades indicate mixed graphidoid and thelotremoid taxa. Bootstrap support is indicated for major clades and figures with detailed clade information are indicated for each clade. The entire, detailed tree is available as Appendix 2.
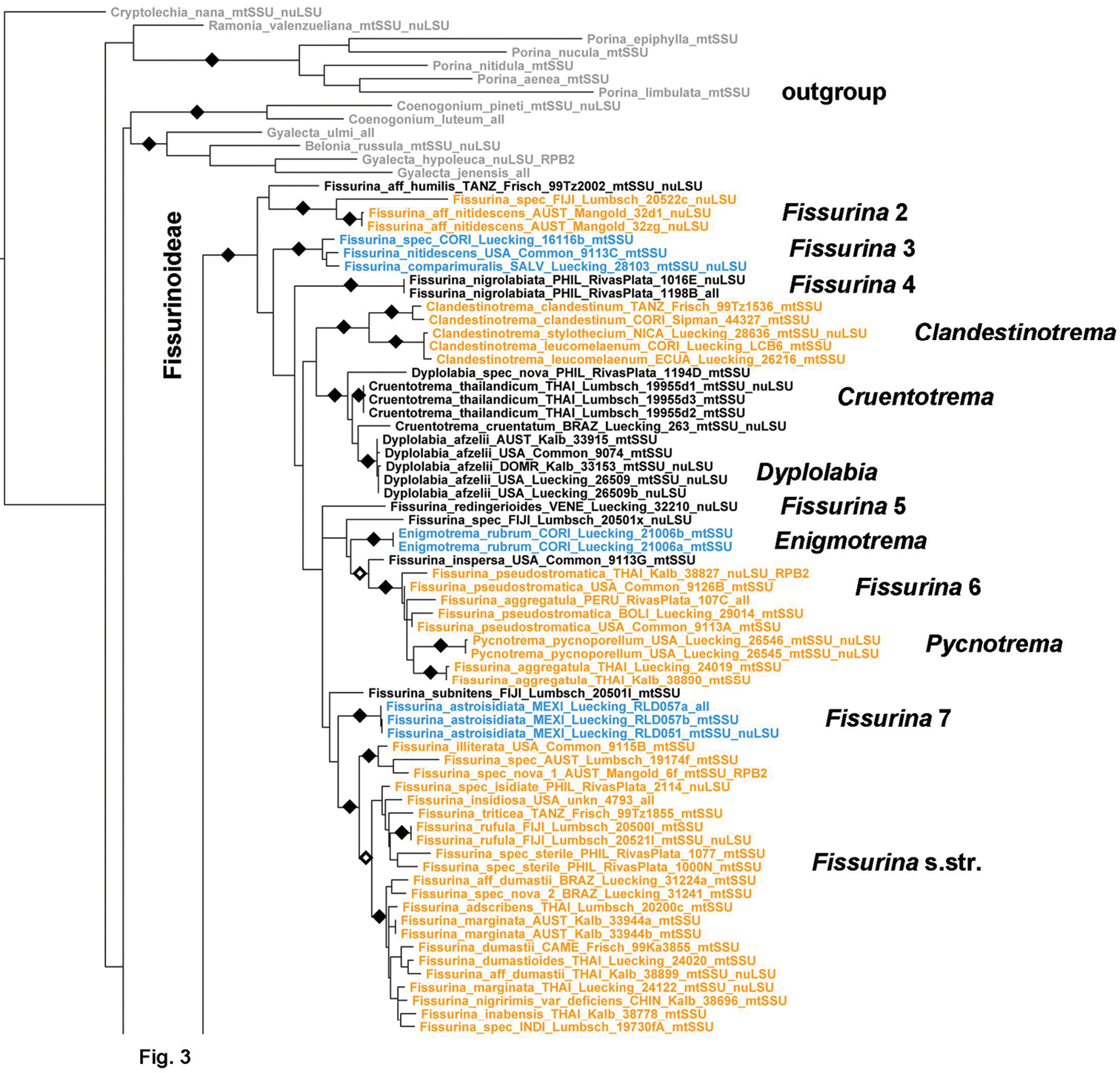
Within subfamily Fissurinoideae, species of Fissurina s. lat. form several clades, indicating that the genus as currently defined is polyphyletic (Fig. 2). Most of the clades correspond to particular morphotypes: the Fissurina humilis, Fissurina nitidescens, and Fissurina nigrolabiata clades (Fissurina 2–4) represent species with carbonized lirellae, the Fissurina pseudostromatica clade (Fissurina 6) species with pseudostromatic lirellae, the Fissurina astroisidiata clade (Fissurina 7) a species with platythecioid lirellae, and the Fissurina dumastii clade (Fissurina s. str.) species with uncarbonized lirellae. Nested within the backbone are the clades corresponding to the genera Clandestinotrema, Cruentotrema, Dyplolabia, Enigmotrema, and Pycnotrema. Clandestinotrema forms two sister clades, one corresponding to species with narrow apothecial pores with entire margin and finger-like columella, represented by the type species, Clandestinotrema clandestinum, and the other to species with broadly open apothecia with fissured margin and broad-stump-shaped columella, represented by Clandestinotrema leucomelaenum and Clandestinotrema stylothecium. Cruentotrema is nested within Dyplolabia, and each genus forms two separate clades each, which contradicts their pronounced morphological differences. Enigmotrema and Pycnotrema are nested within Fissurina s. lat.
Figure 2.
Detailed topology of the Fissurina clade (subfamily Fissurinoideae). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
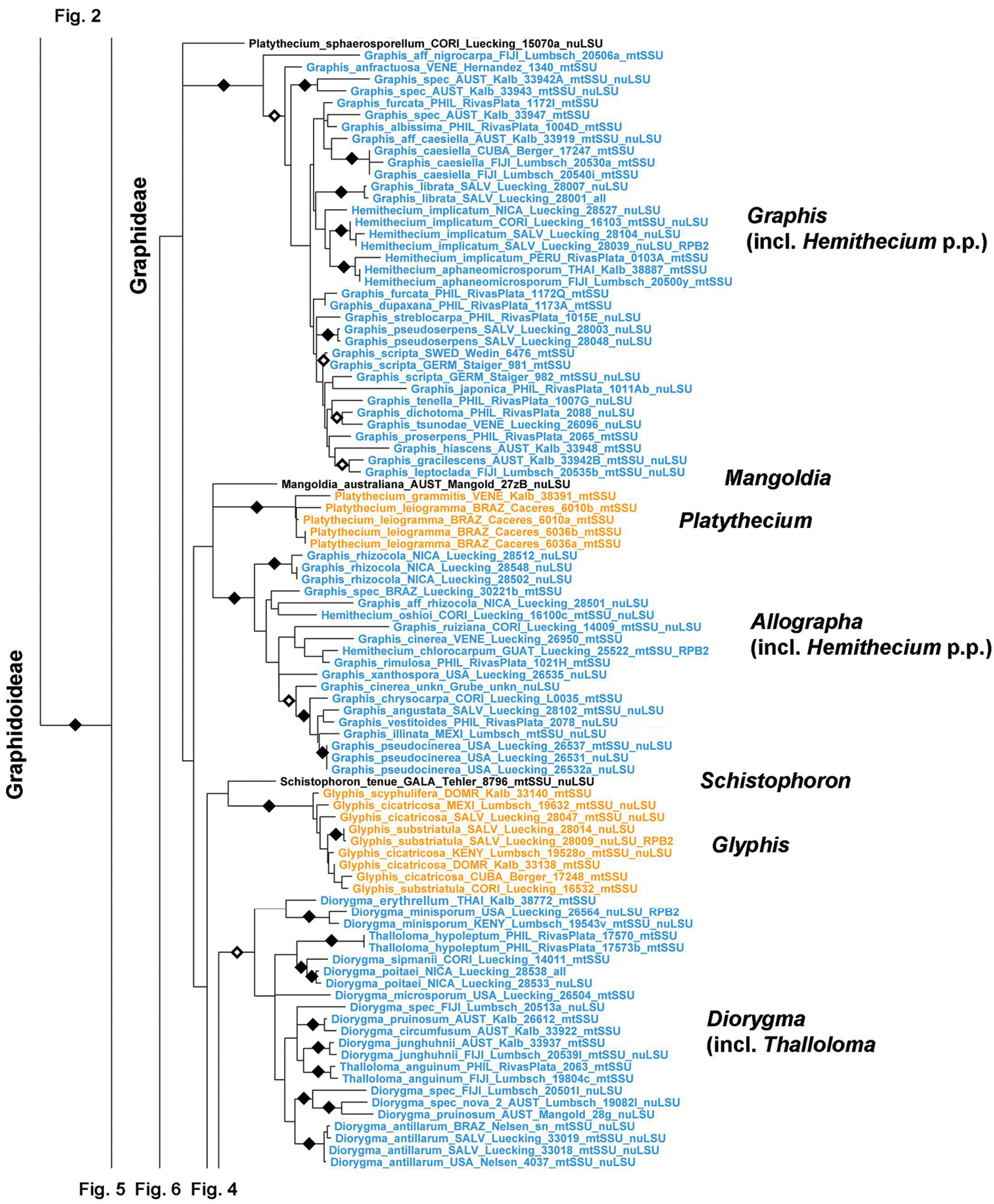
The first clade in subfamily Graphidoideae represents the large tribe Graphideae (unsupported here but supported in previous studies), which can be divided into several larger and smaller clades (Figs 3–5). Graphis s.str., the type genus of the family, is the most unique clade phylogenetically (Fig. 3); it includes part of Hemithecium which corresponds to Graphis except in the uncarbonized lirellae. Platythecium is polyphyletic, forming two separate clades representing the Platythecium allosporellum (Platythecium sphaerosporellum) and Platythecium grammitis groups. Both differ morphologically and chemically in that the first group includes species with basally carbonized excipulum and the testacein chemosyndrome. The genus Allographa includes species of the Graphis cinerea group and part of Hemithecium with large ascospores. A new small genus, Mangoldia (described in a separate paper; Lücking et al. 2012), which is characterized by Phaeographis lecanographa-like ascomata but Graphis-type ascospores, forms a separate clade with unresolved position. Glyphis is strongly supported as monophyletic and the mazaediate Schistophoron persistently clusters at the base of Glyphis but without support. Diorygma includes the genus Thalloloma and also a species formerly placed in the arthonioid genus Herpothallon but now recombined as Diorygma antillarum (Nelsen et al. 2012).
Figure 3.
Detailed topology of the Graphis, Platythecium, Allographa, Glyphis, and Diorygma clades (subfamily Graphidoideae tribe Graphideae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
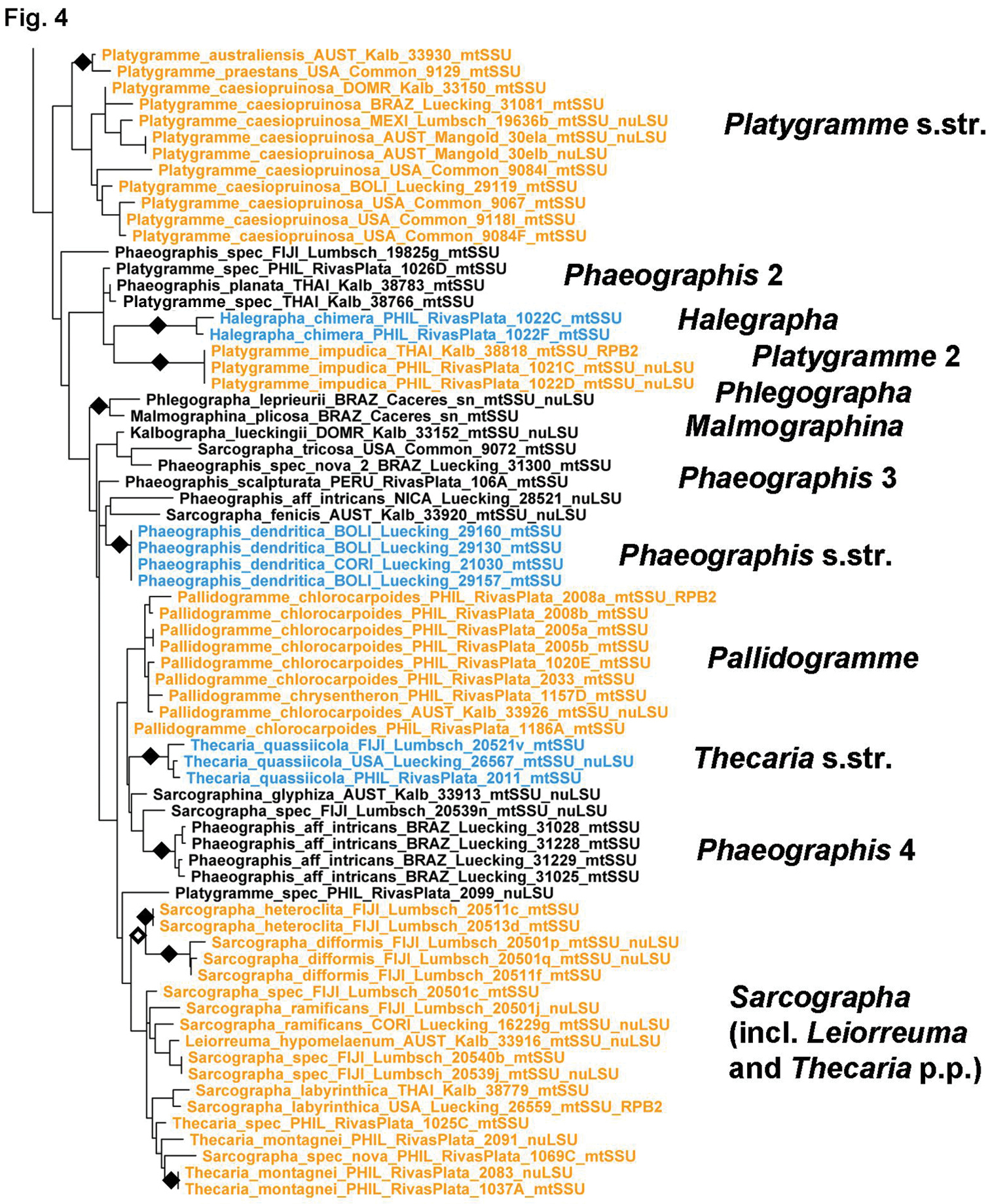
Figure 4.
Detailed topology of the Phaeographis s.lat. clade p.p. (subfamily Graphidoideae tribe Graphideae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
Figure 5.
Detailed topology of the Phaeographis s.lat. clade p.p. (subfamily Graphidoideae tribe Graphideae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
Phaeographis and allied genera (Creographa, Halegrapha, Leiorreuma, Malmographina, Pallidogramme, Phlegographa, Platygramme, Sarcographa, Thecaria) form a complex clade in which generic relationships appear diffuse, with many genera as currently defined being non-monophyletic (Figs 4–5). The Phaeographis lobata group, including Phaeographis lecanographa and allies, forms an unsupported sister-group relationship to the remaining species (Fig. 4). The clade itself is supported by both molecular data and morphological characters and deserves generic status. Species of the Phaeographis intricans group from a strongly supported clade sister to all remaining species, for which the name Creographa is available; however, one species with identical morphology, anatomy, and chemistry, P. aff. intricans (Phaeographis 4), clusters outside this clade, suggesting a remarkable case of parallel evolution. Platygramme s.str. appears to be monophyletic but some species currently classified in that genus, with a different lirellae morphology (thick labia and concealed disc), such as Platygramme impucida (Platygramme 2) cluster outside (Fig. 5). Halegrapha is supported as monophyletic, with an unsupported sister-group relationship to Platygramme impudica (Platygramme 2). Both share Graphis-like lirellae but differ in thallus morphology. Two morphologically unique species classified in separate genera, Malmographina plicosa and Phlegographa leprieurii, form a supported sister group, unsupported sister to the remaining species of Phaeographis s.lat. Malmographina resembles Allographa in lirellae morphology, whereas Phlegographa is externally similar to Glyphis. The remaining species form several separate clades representing Phaeographis s.str., Phaeographis s.lat. (Phaeographis 2–4), Pallidogramme, Sarcographa, and Thecaria. The latter is polyphyletic, with Thecaria montagnei nested within Sarcographa; also nested within that genus is Leiorreuma hypomelaenum, whereas Sarcographina glyphiza falls outside.
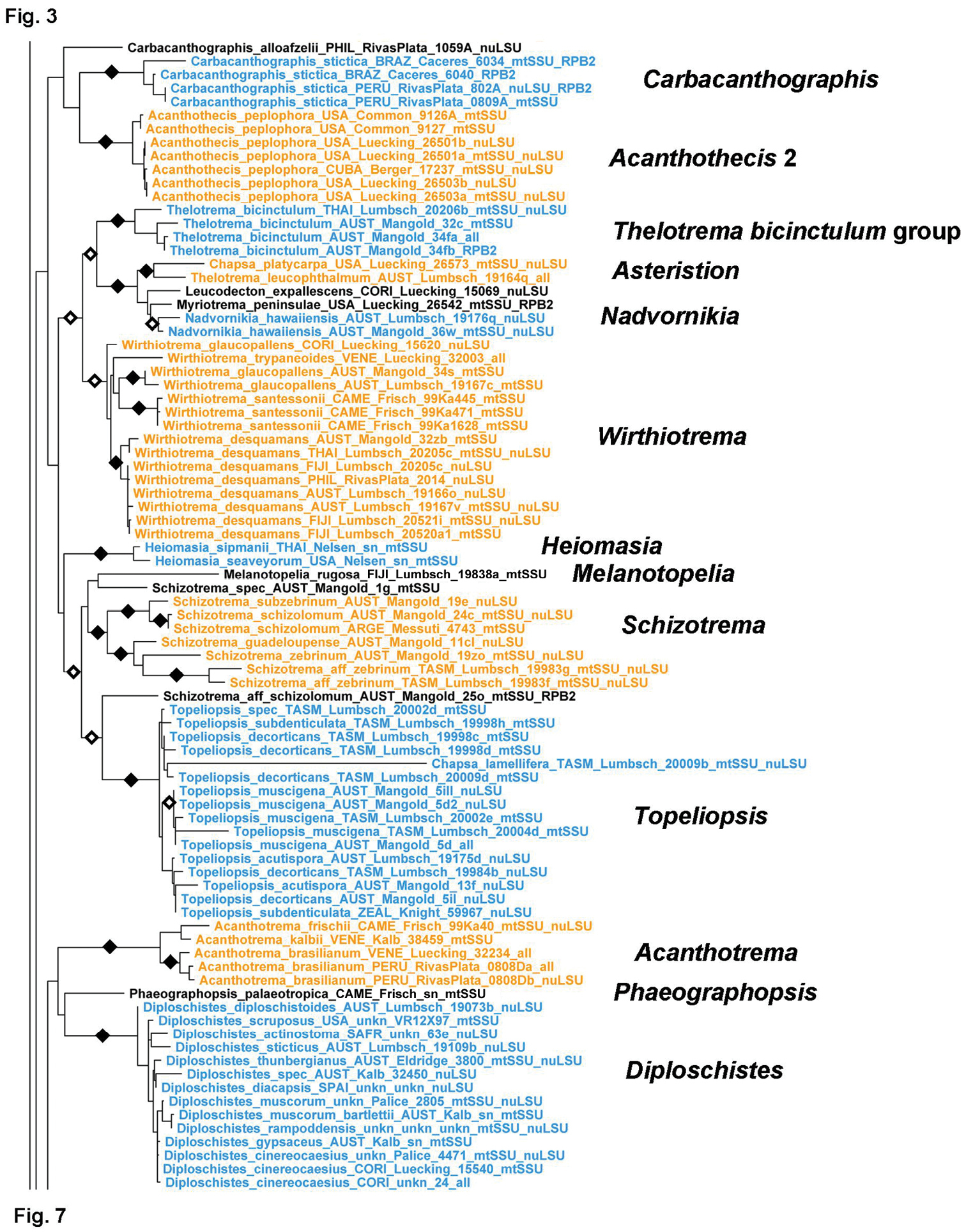
The next clades in subfamily Graphidoideae (Fig. 6) are small clades that were previously assigned to tribe Thelotremateae (Rivas Plata et al. 2012a) but in this study are not supported to form part of that tribe; their phylogenetic position remains unresolved. The Carbacanthographis clade includes two groups with lirellate ascomata that have apically spinulose periphysoids (Carbacanthographis) or paraphyses (Acanthothecis 2); the two lineages are otherwise separated by carbonized (Carbacanthographis) versus uncarbonized (Acanthothecis 2) lirellae, and the latter also differ in their corticate thallus and striate lirellae. This clade is phylogenetically and morphologically quite distinct from Acanthothecis s.str. (see below; Fig. 9) and deserves generic status. The Wirthiotrema clade includes genera and species previously mostly classified as Thelotrema, but apparently not related to Thelotrema s.str. The dominant secondary chemistry of this clade is stictic acid, present in nearly all species. Wirthiotrema itself is monophyletic and characterized by a dense, splitting cortex and non-amyloid ascospores with thickened septa. A second subclade is formed by Thelotrema bicinctulum, which resembles a Myriotrema with double margin but differs in its chemistry and rudimentary periphysoids. A third subclade is formed by Chapsa platycarpa and Thelotrema leucophthalmum, two species that, although previously assigned to different genera, are remarkably similar morphologically. The name Asteristion is available for this clade. Finally, the Nadvornikia subclade includes the mazaediate genus Nadvornikia as well as two non-mazaediate, lepadinoid species, Leucodecton expallescens and Myriotrema peninsulae. The latter is remarkably similar to Nadvornikia superficially except for the persistent, non-mazaediate hymenium. The next clade comprises the enigmatic, sterile genus Heiomasia. The next clade is formed by Topeliopsis and allied genera (Melanotopelia and Schizotrema), the genus Schizotrema apparently not being monophyletic, with one undescribed species forming a sister group relationship with Topeliopsis and another with Melanotopelia. Chapsa lamellifera is supported as nested within Topeliopsis but on a very long branch; morphologically it fits into that genus but differs by its very large apothecia. Finally, the last, unsupported clade comprises three subclades corresponding to three genera with unique morpho-ecological features: the chroodiscoid Acanthotrema, the mazaediate-lirellate Phaeographopsis, and the chroodiscoid-lecanoroid Diploschistes, which features a chlorococcoid photobiont and usually grows on soil or rock. These genera are morphologically and ecologically very disparate and their clustering might not reflect their true phylogenetic relationships, particularly since a long branch leads to each genus. Diploschistes appears non-monophyletic since Diploschistes ocellatus clusters outside the clade (see below; Fig. 10).
Figure 6.
Detailed topology of the Acanthothecis s. lat., Acanthotrema, Carbacanthographis, Diploschistes, Heiomasia, Nadvornikia, Phaeographopsis, Schizotrema, Topeliopsis, and Wirthiotrema clades (subfamily Graphidoideae). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
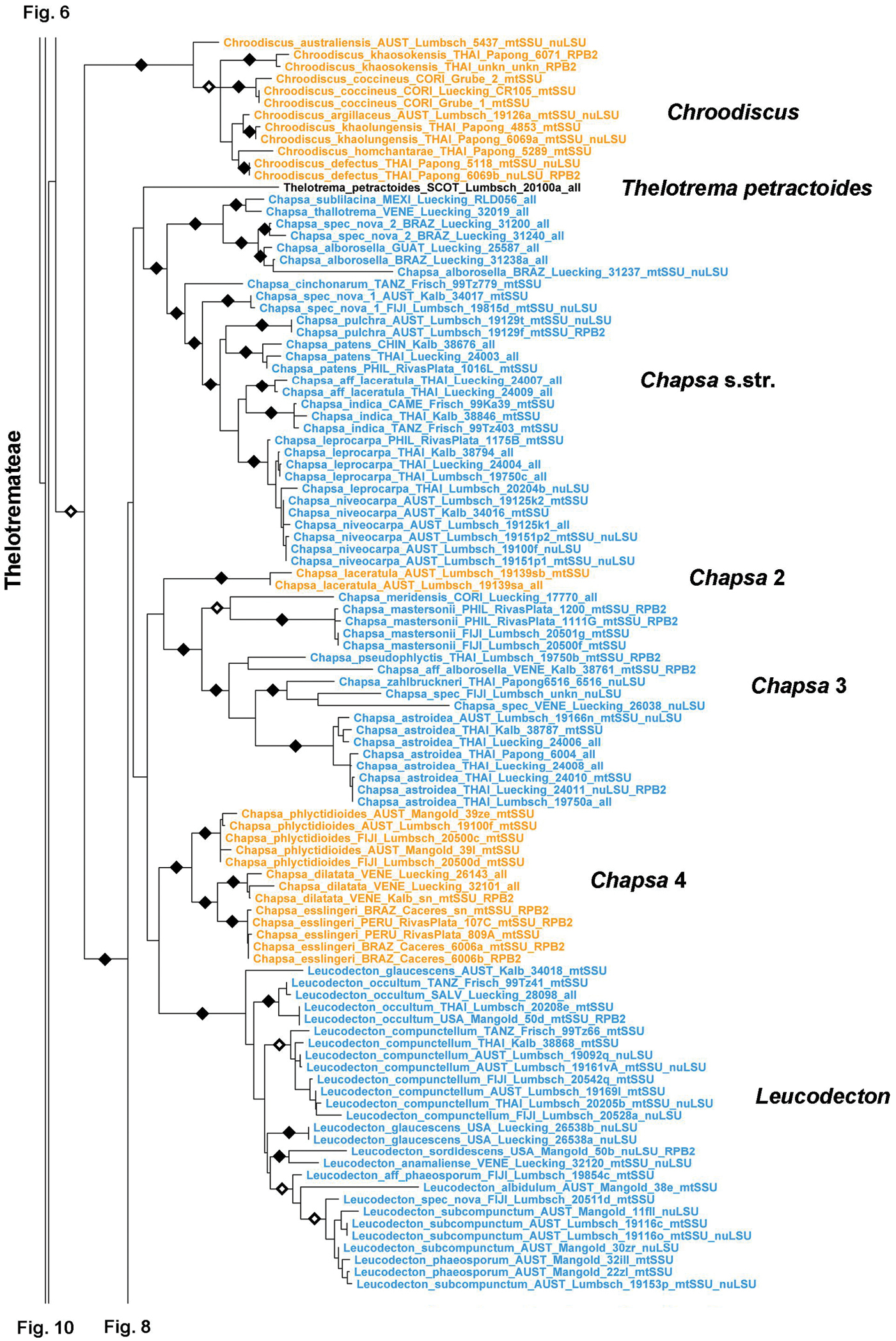
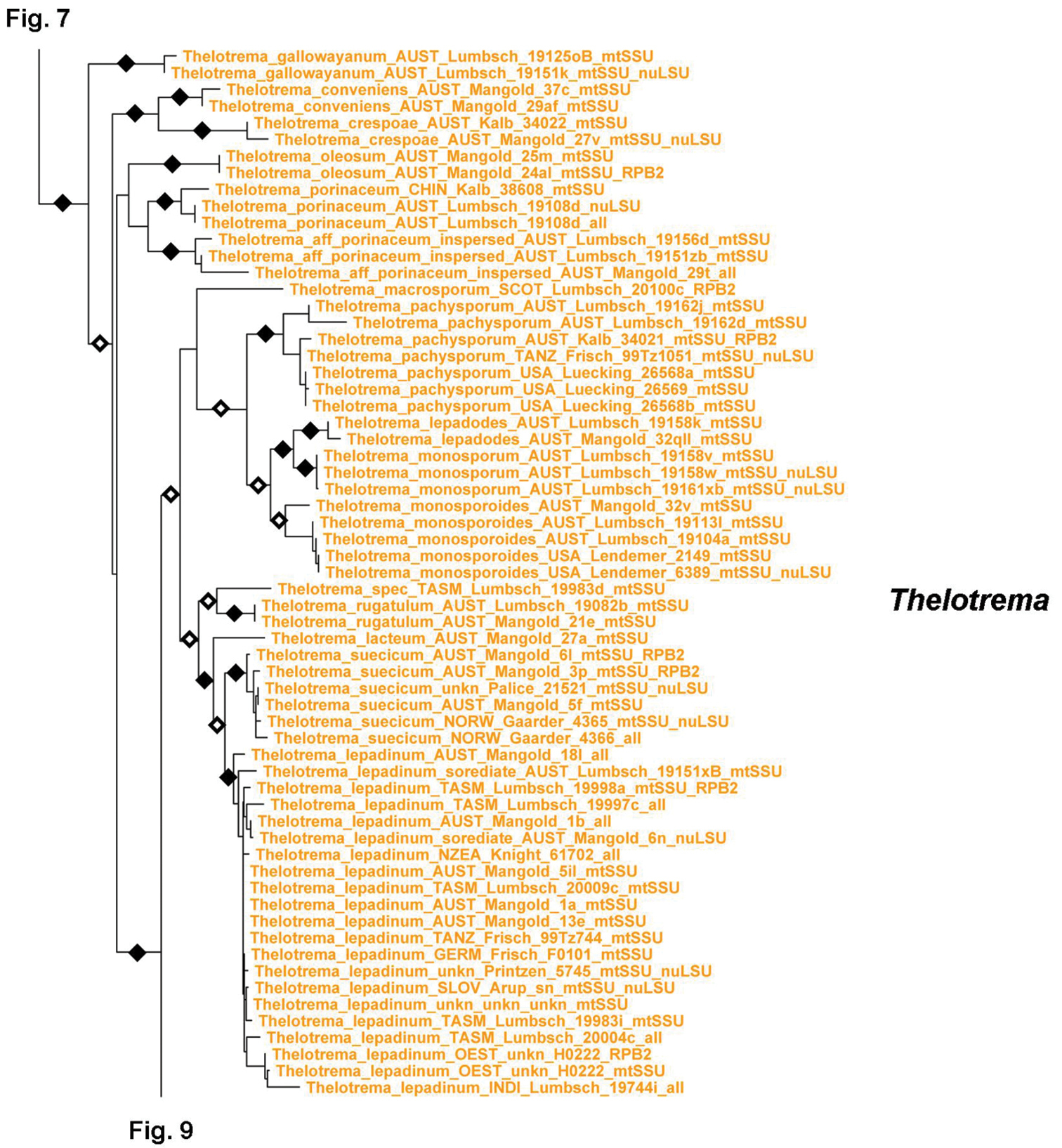
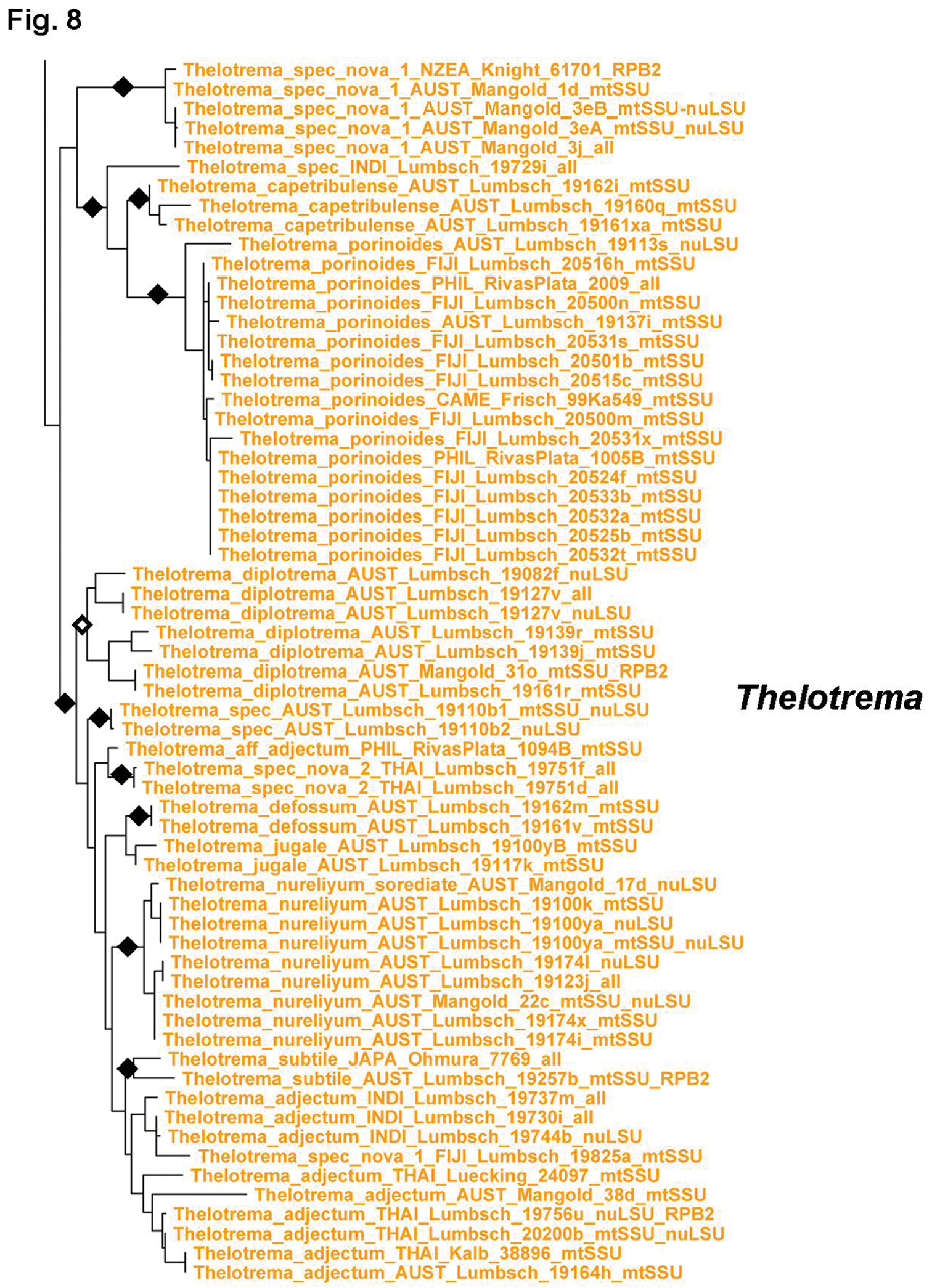
Tribe Thelotremateae forms a large, monophyletic, well-supported clade including the genera Chapsa, Chroodiscus, Leucodecton, and Thelotrema (Figs 7–9). The latter three are monophyletic except Thelotrema petractoides, whereas Chapsa s.lat. is polyphyletic and can be divided into four clades (Fig. 7) depending on the criteria used to recognize each clade (branch length and support): Chapsa s.str., including the type species, Chapsa indica; the Chapsa laceratula clade (Chapsa 2), with Topeliopsis-like apothecia, the Chapsa astroidea clade (Chapsa 3), and the Chapsa dilatata clade (Chapsa 4). Species of Chapsa s.lat. also appear in other clades, such as Chapsa platycarpa in the Wirthiotrema clade (see above; Fig. 6) and Chapsa leprieurii in tribe Ocellularieae (see below; Fig. 12). Thelotrema s.str. is currently the most well-represented clade in terms of sequenced species and geographic cover (Figs 8–9); it shows a well-resolved phylogenetic structure, with Thelotrema gallowayanum, Thelotrema conveniens, Thelotrema oleosum and Thelotrema porinaceum forming supported basal branches in the genus (Fig. 8). All have very narrow pores with the ascomata resembling those of Leucodecton compunctellum and certain Ocellularia s.lat. species (e.g. Ocellularia profunda). The Thelotrema monosporum and Thelotrema lepadinum groups form a supported sister group relationship, which is remarkable since both differ markedly in morphological and anatomical features: ecorticate, white thalli and brown ascospores in the Thelotrema monosporum group versus corticate, yellowish thalli and colorless ascospores in the Thelotrema lepadinum group. These two clades also support the concept of sporomorphs, species with identical morphology that differ in ascospore type only, since the species in the Thelotrema monosporum group are morphologically indistinguishable and also Thelotrema suecicum (small, transversely septate ascospores) and Thelotrema lepadinum (large, muriform ascospores) agree in external morphology. Species separated chiefly by their chemistry, such as Thelotrema porinoides and Thelotrema diplotrema, are supported as separate clades (Fig. 9). The same applies to species with clear versus inspersed hymenium, such as Thelotrema porinoides versus Thelotrema aff. porinoides (Fig. 8), suggesting that secondary chemistry and hymenium inspersion are important species-level characters even if not accompanied by other morphological differences.
Figure 7.
Detailed topology of the Chroodiscus, Chapsa s.lat., and Leucodecton clades (subfamily Graphidoideae tribe Thelotremateae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
Figure 8.
Detailed topology of the Thelotrema clade p.p. (subfamily Graphidoideae tribe Thelotremateae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
Figure 9.
Detailed topology of the Thelotrema clade p.p. (subfamily Graphidoideae tribe Thelotremateae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
The next three clades (Fig. 10) are partially unsupported and their position within the backbone of the family is unresolved. Diploschistes ocellatus, which differs from Diploschistes s.str. in chemistry (norstictic acid) and morphology (apothecia lecanoroid) is here not supported as part of the latter genus (see above; Fig. 6). The Leptotrema clade, which includes the genera Leptotrema and Reimnitzia, had been previously assigned to tribe Ocellularieae (Rivas Plata et al. 2012a) but is here not supported as part of that tribe, although it usually clusters at the base of the latter. The Acanthothecis clade, including the type species, Acanthothecis hololeucoides, is not well-supported and also includes two species previously assigned to Topeliopsis but with a morphology similar to Acanthothecis, Topeliopsis darlingtonii and Topeliopsis elixii. This clade requires further study.
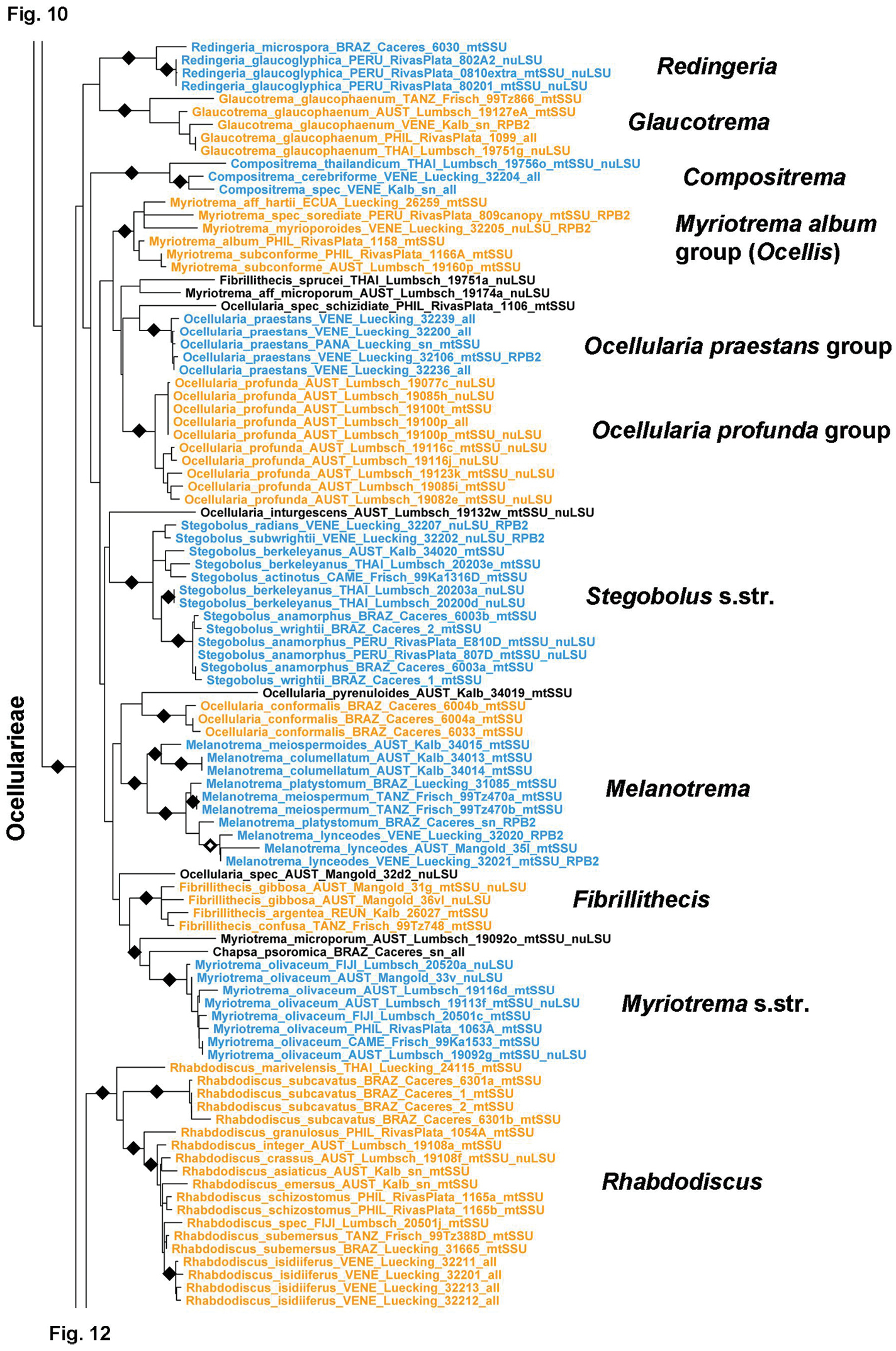
Figure 10.
Detailed topology of the Diploschistes ocellatus, Leptotrema, and Phaeographopsis clades (subfamily Graphidoideae). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
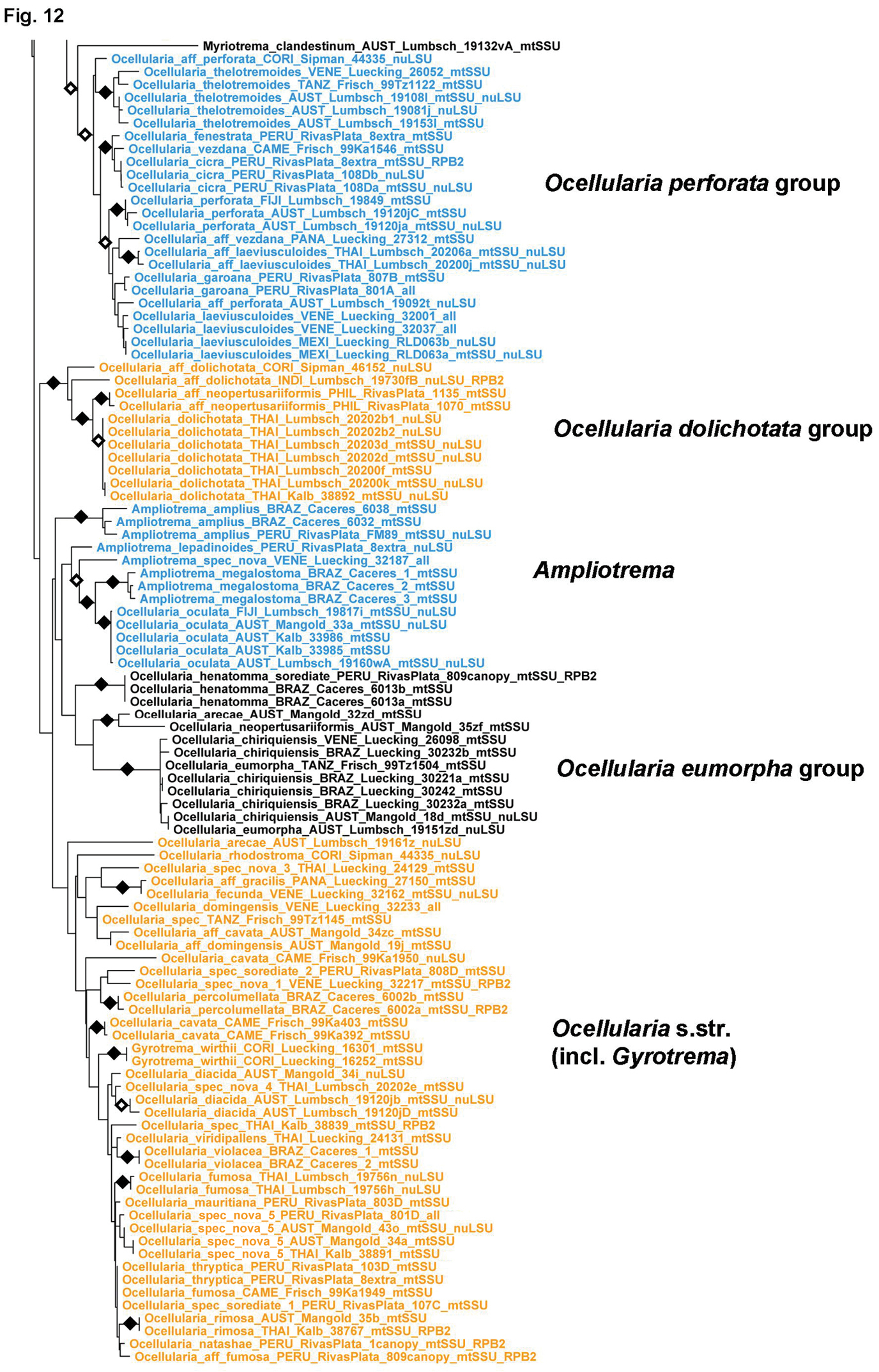
The last clade represents the large tribe Ocellularieae (Figs 11–13). It includes most of the species traditionally assigned to Myriotrema and Ocellularia but apparently corresponds to a much larger number of genus-level clades, some of them already recognized as Fibrillithecis, Melanotrema, Redingeria, Rhabdodiscus, and Stegobolus (Frisch et al. 2006) and others, such as Compositrema, Glaucotrema, and Rhabdodiscus, established recently as part of a detailed study of this clade (Rivas Plata et al. 2012b). The residual Myriotrema and Ocellularia as defined by Frisch et al. (2006) are still highly polyphyletic. Myriotrema can be divided into Myriotrema s.str., the Myriotrema album group (for which the name Ocellis is available), and the Myriotrema glaucophaenum group (recently separated as Glaucotrema). Ocellularia forms a large clade here named Ocellularia s.lat. (Figs 12–13) but also several smaller clades more closely related to Myriotrema s.lat., such as Ocellularia conformalis, Ocellularia inturgescens, Ocellularia praestans, Ocellularia profunda, and Ocellularia pyrenuloides (Fig. 11). Stegobolus s.lat. is divided into the distantly related clades Stegobolus s.str. (ascomata uncarbonized with thick, fuzzy proper margin) and Rhabdodiscus (ascomata carbonized with thin, compact and smooth proper margin), the latter forming an unsupported sister to Ocellularia s.lat. (Fig. 11). The Ocellularia s.lat. clade is highly structured, including several strongly supported subclades with homogeneous morpho-chemical features (Figs 12–13): the Ocellularia terebrata group with psoromic acid and olive-green thalli; the Ocellularia wirthii group with psoromic acid and white thalli; the Ocellularia baileyi group with nornotatic acid; the Ocellularia microstoma group (for which the name Macropyrenium is available) with large, annulate ascomata and large, muriform ascospores, but variable chemistry; the Ocellularia bahiana group (for which the name Stigmagora is available) with often grainy thalli due to columnar clusters of crystals, an often irregular pseudocolumella, and protocetraric acid as predominant secondary compound; the Ocellularia perforata group with small, often myriotremoid ascomata and psoromic and protocetraric acid or no substances; the Ocellularia dolichotata group, lacking substances but with large, transversely septate ascospores; and the Ocellularia eumorpha group, with large ascomata and ascospores and hypoprotocetraric acid. Ampliotrema, with ecolumellate, often pigmented ascomata, inspersed hymenium, and protocetraric acid, appears as a paraphyletic residual basal to the Ocellularia eumorpha clade and Ocellularia s.str. The latter is unsupported but always monophyletic and chiefly includes species with pigmented medulla and hypoprotocetraric or nornotatic acid or cinchonarum or other unknowns. Gyrotrema is nested within Ocellularia s.str., although its chroodiscoid ascomata are quite different from the ocellularioid ascomata of the other species. Another gyrotremoid-stegoboloid species, Ocellularia percolumellata, is also nested in this clade. Perhaps the most unexpected surprise of this study is the supported placement of Chapsa leprieurii within Ocellularia s.lat.; except for the corticate, olive-brown thallus, this species agrees with Chapsa s.lat. in ascomata features.
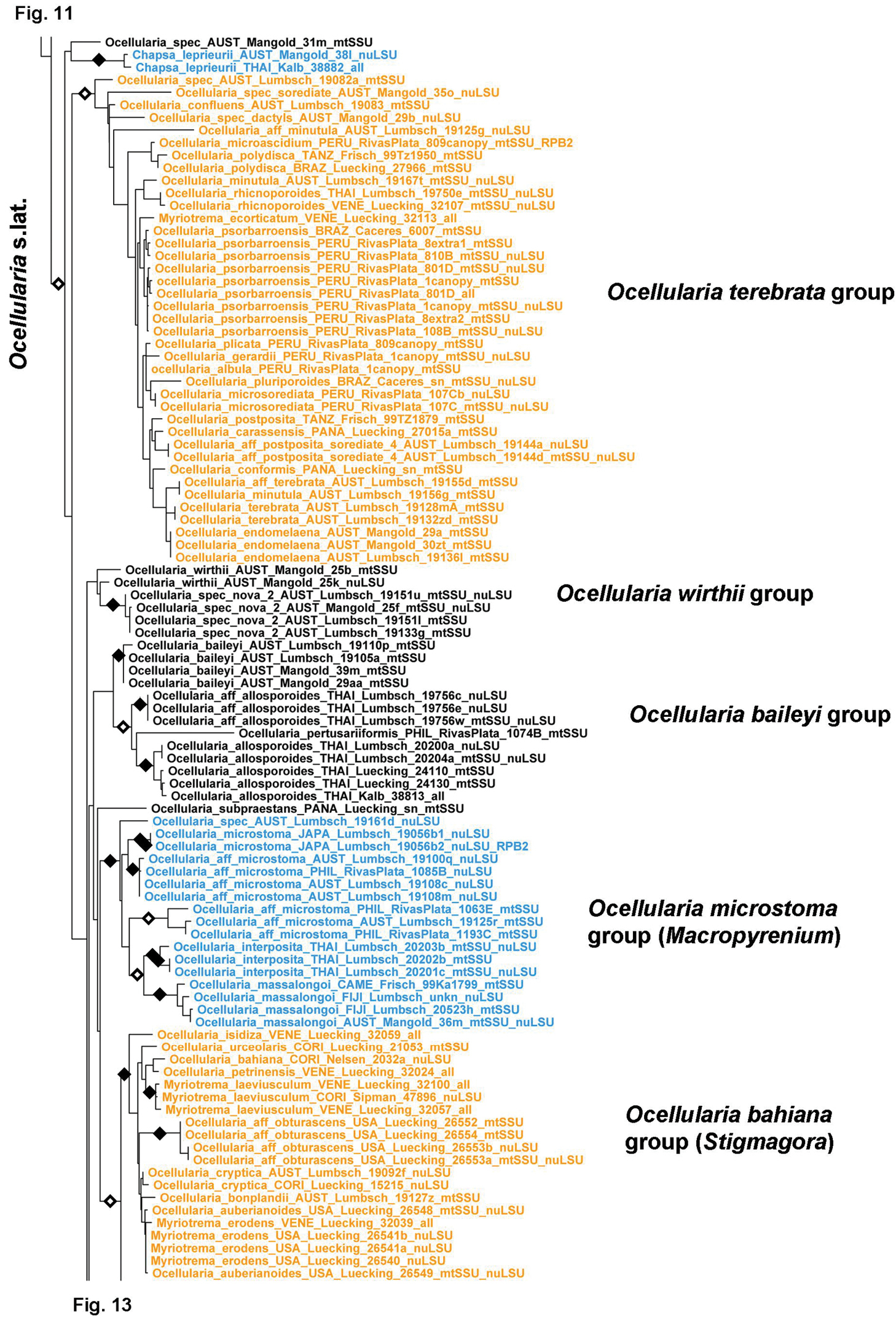
Figure 11.
Detailed topology of the Myriotrema s.lat. clade and relatives (subfamily Graphidoideae tribe Ocellularieae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
Figure 12.
Detailed topology of the Ocellularia s.lat. clade p.p. (subfamily Graphidoideae tribe Ocellularieae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
Figure 13.
Detailed topology of the Ocellularia s.lat. clade p.p. (subfamily Graphidoideae tribe Ocellularieae p.p.). Genus level clades are highlighted. Bootstrap support is indicated as black (90% and higher) and black-and-white (70% and higher) symbols. For specimen information and GenBank numbers see Appendix 1. The entire, fully resolved tree with detailed bootstrap support values is available as Appendix 2.
Of the 428 species included in this analysis, 185 (42%) are represented by two or more (up to 21 in the case of Thelotrema lepadinum) OTUs (total of 658 out of 922 OTUs), providing a base for a first test of species circumscriptions. The identifications used in this analysis are based on critical cross-examination of all available material including comparison to authentic type specimens, meaning that specimens bearing the same name were found to be morphologically identical. Of the 185 species with more than one sample, 148 (80%) were monophyletic; 20 were paraphyletic, and 17 were polyphyletic. There are several reasons to explain species-level paraphyly, such as insufficient resolution in closely related species complexes, especially in such a large phylogeny where terminal clades suffer from the effect of gap-rich alignment portions. Also, incomplete lineage sorting could cause paraphyly. Polyphyly can be explained by incomplete and non-overlapping gene sampling between OTUs of the same species, as well as incorrect species concepts suggesting (semi-)cryptic species. In the present case, two cases of paraphyly (Hemithecium implicatum, Ocellularia wirthii) and eleven cases of polyphyly (Diorygma pruinosum, Glyphis substriatula, Leucodecton glaucescens, Melanotrema platystomum, Ocellularia aff. microstoma, Ocellularia arecae, Ocellularia cavata, Ocellularia fumosa, Stegobolus berkeleyanus, Topeliopsis subdenticulata, Wirthiotrema glaucopallens) can be explained by incomplete gene sampling, e.g. mtSSU available for one OTU of a species and nuLSU for another, but not both for the same OTU. Another 12 cases of paraphyly (Chapsa leprocarpa, Clandestinotrema leucomelaenum, Fibrillithecis gibbosa, Leucodecton phaeosporum, Leucodecton subcompunctum, Myriotrema erodens, Ocellularia auberianoides, Ocellularia chiriquiensis, Ocellularia eumorpha, Platythecium leiogramma, Stegobolus anamorphus, Stegobolus wrightii) are potentially explained by insufficient resolution at the terminal clade level or incomplete lineage sorting or a combination of both. This leaves six cases of paraphyly (Fissurina pseudostromatica, Ocellularia diacida, Pallidogramme chlorocarpoides, Sarcographa ramificans, Thecaria montagnei, Thelotrema adjectum) and another six of polyphyly (Fissurina aggregatula, Fissurina marginata, Glyphis cicatricosa, Graphis scripta, Topeliopsis acutispora, Topeliopsis decorticans) that cannot be explained by these causes and thus appear to indicate issues with current species circumscriptions.
The species level approach also allowed for testing the existence of widely distributed, intercontinental, pantropical, and cosmopolitan species. Of the 148 monophyletically resolved species, based on the available molecular data, two were confirmed as cosmopolitan taxa (the closely related sister species Thelotrema lepadinum and Thelotrema suecicum), 20 as pantropical (Dyplolabia afzelii, Glaucotrema glaucophaenum, Graphis caesiella, Leucodecton occultum, Melanotrema lynceodes, Ocellularia albocincta, Ocellularia cryptica, Ocellularia rhicnoporoides, Ocellularia thelotremoides, Phaeographis alcicornis, Phaeographis intricans, Phaeographis lecanographa, Phaeographis lobata, Phaeographis platycarpa, Platygramme caesiopruinosa, Reimnitzia santensis, Sarcographa labyrinthica, Thecaria quassiicola, Thelotrema monosporoides, Thelotrema pachysporum), four as at least Gondwanan (Neotropics and tropical Africa) but possibly pantropical (Clandestinotrema clandestinum, Diorygma minisporum, Ocellularia polydisca, Rhabdodiscus subemersus), four as at least paleotropical (tropical Africa, Asia and Australia) but possibly in part pantropical (Chapsa indica, Leucodecton compunctellum, Myriotrema olivaceum, Ocellularia massalongoi), and one austral (Argentina and Australia: Schizotrema schizolomum).
Discussion
The phylogeny presented here is the most comprehensive analysis of Graphidaceae known to date compared to previous studies (Staiger et al. 2006, Mangold et al. 2008a, Rivas Plata and Lumbsch 2011). The taxon sampling, with 428 species and 908 ingroup OTUs, represents about 25% of the currently accepted species and hence provides substantial insight into the phylogenetic relationships of major clades, genera, and species. It is obvious that the evolution within this family is much more complex than reflected by previous classifications which were in use until about ten years ago (Wirth and Hale 1963, 1978; Hale 1974, 1978, 1980, 1981; Archer 1999, 2000, 2001a–d, 2002). Many of the relationships found based on molecular data would have been impossible to predict using morphological characters, such as the split into subfamilies Fissurinoideae and Graphidoideae or the polyphyly of the large genera Chapsa sensu Frisch et al. (2006), Graphis sensu Staiger (2002) and Ocellularia sensu Frisch et al. (2006a). Also, the inclusion of myriotremoid and ocellularioid lineages, such as Clandestinotrema, Cruentotrema, and Pycnotrema, within Fissurina s.lat., or the separation of morphologically similar taxa like Leptotrema wightii, Leucodecton phaeosporum, and Myriotrema laeviusculum, in unrelated lineages came quite unexpected (Rivas Plata and Lumbsch 2011, Rivas Plata et al. 2012a). Another novel clade detected in the present study is the Nadvornikia-Wirthiotrema clade, which contains the mazaedioid Nadvornikia amidst an assemblage of Thelotrema-like species that are unrelated to Thelotrema s.str. While apparently most morphological characters such as ascomata shape, excipular carbonization, and the presence of a columella, evolved multiple times within the family in distantly related clades (Rivas Plata and Lumbsch 2011), surprisingly one of the most conserved character complexes turned out to be secondary chemistry. Most clades have a predominance of particular chemosyndromes, such as stictic and norstictic acids in the tribes Graphideae and Thelotremateae and various smaller clades falling outside these tribes, as well as psoromic, protocetraric, and hypoprotocetraric acids, and cinchonarum unknowns, in tribe Ocellularieae.
The most controversially discussed higher-level changes in the classification of Graphidaceae were the inclusion of Thelotremataceae, Gomphillaceae, and Asterothyriaceae (= Solorinellaceae) in this family (Aptroot 2012; Hodkinson 2012, Rivas Plata et al. 2012a; Sipman et al. 2012). The non-monophyly of Graphidaceae versus Thelotremataceae was already indicated in early phylogenetic studies (Staiger et al. 2006) and later confirmed by Mangold et al. (2008a). The current, much larger dataset clearly supports the conclusion that Graphidaceae and Thelotremataceae cannot be separated at the family level. In order to maintain the name Thelotremataceae, it would have to be restricted to tribe Thelotremateae, which would require to establish several separate families for tribes Graphideae and Ocellularieae, as well as for a number of smaller clades. Since the only argument separating the two families historically was ascomata shape (round in Thelotremataceae versus lirellate or pseudostromatic in Graphidaceae), a classification restricting the name Thelotremataceae to tribe Thelotremateae would be misleading and would be an effort to maintain a well-known name, rather than reflecting a scientific advance. In the light of multiple evolution of ascomata types in different clades, merging the two families and instead delimiting infrafamily-level clades reflects the phylogenetic relationships much better.
A more difficult issue is the inclusion of Gomphillaceae and Asterothyriaceae within Graphidaceae. At first glance, this appears to be a purely phylogenetic requirement when applying the concept of strict monophyly. According to Aptroot (2012), Graphidaceae could technically be maintained as paraphyletic residual giving origin to a monophyletic Gomphillaceae (including Asterothyriaceae and Solorinellaceae). The problem of paraphyly is controversely discussed and no widely accepted solution has been proposed (Podani 2010, Hörandl and Stuessy 2010). However, the topology of the three clades proposed as subfamilies of Graphidaceae, Graphidoideae, Fissurinoideae, and Gomphilloideae (Rivas Plata et al. 2012a) does not reflect what is widely understood as paraphyly: a comb-shaped backbone from which individual clades emerge in different positions. On the contrary, the three subfamilies form three strongly supported clades on long stem branches each, and multigene studies show strong support for Fissurinoideae and Gomphilloideae being sister to each other and together sister to Graphidoideae (Baloch et al. 2010, Rivas Plata et al. 2012a, b, Nelsen et al. 2012). Therefore, splitting Fissurinoideae and the more closely related Gomphilloidae at the family level while keeping Fissurinoideae and the more distantly related Graphidoideae in a single family Graphidaceae would not create a paraphyletic taxon but would strongly disagree with the phylogenetic relationships of these taxa. Another alternative would be the recognition of three families, Gomphillaceae, Fissurinaceae, and Graphidaceae, as suggested by Hodkinson (2012). However, such a classification would not actually solve the dilemma of the apparent discrepancy between molecular phylogeny and morphological character evolution, since it would separate two clades, Fissurinoideae and Graphidoideae, at the family level with virtually no phenotypic character supporting such a separation.
One criterion that could be used to compare the alternative classifications is information content. Obviously, merging all clades into one family has the information content zero at the family level. Therefore, splitting them into three families makes only sense if the information content is greater than zero. This can be tested by using a pairwise comparison: assuming that Gomphilloideae (GO) are morphologically distinct from both Fissurinoideae (FI) and Graphidoideae (GR), whereas the latter two are not distinguishable, the pairwise comparison would result in two comparisons that are informative (GO versus FI and GO versus GR) and one comparison that is not informative (FI versus GR). Setting informative pairwise comparisons to +1 and non-informative to –1, the total would be +1, thus greater than zero, which would favor the 3-family solution partially suggested by Hodkinson (2012) over the 1-family solution suggested by Rivas Plata et al. (2012a). However, the distinction of Gomphillaceae from the other two families is not as straightforward as considered by Hodkinson (2012) and its inclusion within a wider Graphidaceae not as counterintuitive as it appears at first glance (Sipman et al. 2012). Gomphillaceae is largely separated from Graphidaceae by a chlorococcoid versus trentepohlioid photobiont, the anastomosing versus unbranched paraphyses, the thin-walled and non-amyloid versus graphidoid and amyloid ascospores, and the hyphophorous conidiomata, as well as the predominantly foliicolous growth habit (Lücking et al. 2004, Lücking 2008). However, these differences disappear when considering the entire range of variation in both families: Graphidaceae includes taxa with chlorococcoid photobiont (Diploschistes) and species with thin-walled, non-amyloid ascospores (Chroodiscus, Acanthotrema). Other groups, such as Diorygma, Dyplolabia, and Ocellularia s.lat., have at least partially anastomosing paraphyses. Hyphophores are not present in all species of Gomphillaceae and are absent in the genera Asterothyrium, Gyalidea, and Psorotheciopsis (Henssen and Lücking 2002, Lücking 2008). Many Gomphillaceae, particularly in the genera Echinoplaca and Gyalideopsis, are corticolous, and in Graphidaceae, genera such as Chroodiscus are exclusively foliicolous. In addition, both groups share important features, such as the usually zeorine, hemiangiocarpous ascomata with a strong tendency to become lobate-lirellate, and the graphidoid ascus type. Therefore, considering the whole range of variation, there is no clear limit between these two families. Notably, Aulaxina was maintained in Graphidaceae by Santesson (1952) and Asterothyrium at some point was suggested to belong in Thelotremataceae and being related to Chroodiscus (Vezda and Poelt 1987; Aptroot et al. 1994). This view is also supported by the undisputed inclusion of Diploschistes in Graphidaceae, a genus that is ecologically very different from the remaining Graphidaceae and also genetically distinct, and which had been included in a separate family, Diploschistaceae, in the past (Zahlbruckner 1905). Hence we prefer to maintain a larger circumscription of Graphidaceae at this point.
Several genera currently placed in Graphidaceae have not yet been sequenced. These include Amazonotrema, Anomalographis, Anomomorpha, Diaphorographis, Gymnographopsis, Kalbographa, and Sarcographina (Staiger 2002, Lücking 2007, Kalb 2009, Rivas Plata et al. 2012a). The type of Sarcographina is unrelated to the sequenced Sarcographina glyphiza which therefore does not represent that genus. Also, the placement of Kalbographa lueckingii is doubtful since this species is morphologically and chemically closer to Phaeographis dendritica that to Kalbographa s.str. We therefore consider Kalbographa as of uncertain phylogenetic placement. Most of these genera have thin-walled ascospores different from most Graphidaceae but similar to, for example, Phaeographopsis. Since the latter forms a single clade with unresolved position, we also anticipate these unsequenced genera to represent further separate clades within the family. Anomomorpha is expected to be closely related to Platythecium str., since it shares morphological and chemical characteristics with that genus (Staiger 2002).
Clades and taxa: progress and problems
Subfamily Fissurinoideae
This subfamily currently includes six accepted genera. The core genus, Fissurina, includes more than 70 known species (Staiger 2002, Lücking, in prep.) and is highly polyphyletic, as obvious from more than half of the species sequenced. The largest, supported clade is centered around the type species, Fissurina dumastii. Another large clade includes predominantly pseudostromatic species centered around Fissurina pseudostromatica; for this clade, the name Medusulina is potentially available, but more species need to be sequenced to test whether this clade corresponds to pseudostromatic species only. Species with carbonized excipulum form at least three supported clades at the base of the subfamily; they differ chiefly in ascomata morphology: fissurinoid to chroodiscoid in the Fissurina humilis and Fissurina nitidescens clades and graphidoid in the Fissurina nigrolabiata clade. Several other species of Fissurina do not cluster with these larger clades. A few more critical species require sequencing and detailed morpho-anatomical studies are necessary to elucidate whether all these clades can be recognized as separate genera. For instance, species of Fissurina s.lat. partly feature warty paraphyses and also vary in ascospore amyloidity (Staiger 2002) and these could be features distinguishing between clades.
Clandestinotrema is strongly supported as a monophyletic clade, distinguished from other taxa in this subfamily by the whitish, often loosely corticate or ecorticate thallus and ocellularioid, usually carbonized and often columellate ascomata. The taxonomy of this genus is well-resolved (Rivas Plata et al. 2012a, Sipman et al. 2012), but a few more of the currently accepted 12 species need to be sequenced to confirm monophyly of this clade. There is some indication that species with pore-like versus broadly open apothecia form two subclades that each might deserve genus or subgenus rank, but more sequences are required to test this assumption. A peculiar character shared with Cruentotrema, Dyplolabia, and several species of Fissurina, are the astrothelioid ascospores, which also help to distinguish this genus from similar species in Melanotrema and Ocellularia s.lat. (Rivas Plata and Lumbsch 2011).
Cruentotrema and Dyplolabia are two very closely related genera, although morphologically quite disparate, being either chroodiscoid with bright red medullary pigment or in the latter graphidoid with thick white pruina containing lecanoric acid (Kalb and Staiger 2000; Rivas Plata and Lumbsch 2011; Rivas Plata et al. 2012a). Each genus currently contains three species and two species each have been sequenced. In the best-scoring ML tree, Cruentotrema appears nested within Dyplolabia, but without support, and SH testing performed on a subset of the data including the Cruentotrema-Dyplolabia clade and its sister group does not reject the possibility of both genera being monophyletic (results not shown). Considering the morphological differences between the two genera and the infrageneric uniformity within each of them, non-monophyly seems indeed unlikely and we are planning sequence additional loci to elucidate the relationships of these genera. However, this could be an ideal group to test the potential effect of incomplete lineage sorting on incongruence between gene phylogenies and actual taxon-level lineages.
The recently described, monospecific genus Enigmotrema is very similar to Cruentotrema but its apothecia remain closed for a long time and are bright red from the outside, resembling the pyrenocarpous lichen Pyrenula cruenta (Sipman et al. 2012). Inspite of its similarities with Cruentotrema, the genus clusters outside the Cruentotrema-Dyplolabia clade and appears more closely related to the Fissurina pseudostromatica clade. Nested within this clade is another monospecific genus, Pycnotrema, which resembles species of Myriotrema with its small, rounded apothecial pores (Rivas Plata et al. 2012a). Its nested position within the Fissurina pseudostromatica clade needs to be examined further.
Subfamily Graphidoideae: tribe Graphideae
This is the largest clade within the family, with possibly over 600 species. It includes the bulk of the former Graphidaceae with lirellate ascomata, with the exception of Fissurina s.lat., Carbacanthographis, and Acanthothecis. The major clades within this tribe correlate largely with ascospore type (hyaline-amyloid versus brown-hemiamyloid), whereas characters such as excipulum carbonization play a minor role, as shown by the inclusion of species of Hemithecium in either Graphis or Allographa. Thallus morphology also appears to be a good predictor of clade relationships, with white, strongly crystalline thalli characteristic of Graphis and Allographa, ecorticate thalli found mostly in the Diorygma-Thalloloma clade, and olive-green to yellow-brown thalli often lacking crystal clusters in Glyphis and the Phaeographis s.lat. clade. Graphideae are divided into two genetically distinct clades, one corresponding to Graphis s.str. and the other to all other taxa. The genetic uniqueness of Graphis s.str. was only recently established (Rivas Plata et al. 2011, Berger et al. 2011). A large number of species previously included in the revised concept of Graphis sensu Staiger (2002) turned out to belong in a separate genus, Allographa, which is more closely related to the remaining Graphideae. A formal reclassification reflecting this phylogeny has not yet been presented since species with uncarbonized excipulum previously separated in Hemithecium also fall into either Graphis or Allographa and more species of Hemithecium, including its type, need to be sequenced in order to correctly reallocate them. Rivas Plata et al. (2011) already showed that Hemithecium sensu Staiger (2002) was highly polyphyletic and formed at least five unrelated clades. Two of them clustered with either Graphis or Allographa, whereas another two fell into the vicinity of Phaeographis and are currently recognized as Malmographina and Pallidogramme, respectively (Lücking et al. 2008, Cáceres et al. 2012). The fifth clade, consisting of Hemithecium rufopallidum, apparently formed an isolated clade related to Allographa (Rivas Plata et al. 2011). In our current analysis, the species Platythecium grammitis fell into this clade, and re-examination of the material of Hemithecium rufopallidum revealed that our previous identification was incorrect: while this species was present in the voucher material, the sequenced taxon actually belonged to the superficially similar Platythecium leiogramma, thus supporting Platythecium as a separate genus.
The genus Glyphis remains a strongly supported clade within Graphideae, including species with rounded, lirellate, and pseudostromatic ascomata. However, species relationships require further study since the pseudostromatic Glyphis cicatricosa does not appear to be monophyletic. The mazaediate genus Schistophoron, which resembles species of Allographa, consistently comes out as sister to Glyphis, but lacking support. Both genera are morphologically and anatomically quite different. Diorygma and Thalloloma, two genera with similar thallus and ascomata morphology but with different chemical profiles (Staiger 2002, Kalb et al. 2004) appear to be nested within each other and most probably will have to be merged into a single genus. Supposed differences such as hamathecium structure and amyloidity (Staiger 2002, Kalb et al. 2004) do not seem to correlate with the topology found in this clade. On the other hand, Platythecium sensu Staiger (2002) appears to be polyphyletic, corresponding to two morphologically and chemically distinct clades.
The remaining genera all have brown, hemiamyloid ascospores and consistently form a monophyletic clade (Lücking et al. 2011, Rivas Plata et al. 2011, Cáceres et al. 2012), although support was lacking in the present analysis. However, genus-level relationships and genus delimitation within this clade are in need of major revision. Most of the currently accepted genera (Staiger 2002) are para- or polyphyletic at some level, whereas supported monophyletic clades have not been recognized at genus level. Among the latter are the Phaeographis lobata-lecanographa group and the Phaeographis intricans group, for which the name Creographa is potentially available. Platygramme s.str. is monophyletic but its delimitation towards Phaeographis s.lat. is uncertain, since species with distinctly carbonized labia, such as Phaeographis impudica, appear elsewhere in this clade. Most other genera, such as Halegrapha, Malmographina, Pallidogramme, Phlegographa, and Thecaria, show a similar pattern, with the type species and its close relatives forming monophyletic clades but nested within a paraphyletic Phaeographis s.lat. grade. Thecaria montagnei is apparently not close to the type species, Thecaria quassiicola, and the generic name Pliariona is potentially available for this species. Leiorreuma and Sarcographa appear both non-monophyletic, with the core groups forming a single clade in a derived position. Phaeographis s.str. thus far appears to be restricted to the type species, Phaeographis dendritica, which is characterized by its white, ecorticate thallus and non-pruinose ascomata. Most species of Phaeographis s.lat. have a corticate thallus and pruinose ascomata, and the name Ectographis is potentially available for these. However, many more species need to be sequenced in these groups to establish a solid phylogeny and generic concept for Phaeographis and its allies. The most important characters separating clades within this assemblage are ascomata organization (solitary versu pseudostromatic), disc visibility (exposed versus concealed), hymenium inspersion (inspersed versus clear), and excipular and hypothecium carbonization (uncarbonized versus carbonized). On their own, these clades appear to be quite distinctive; however, exactly the same level of variation is found in Graphis and Allographa, two clades that form single genera including between 100 and over 200 species each, and therefore, it would not appear illogical to actually merge Creographa, Ectographis, Halegrapha, Leiorreuma, Malmographina, Pallidogramme, Phaeographis s.str., Phlegographa, Platygramme, Pliariona, Sarcographa, and Thecaria, into a single, large genus Phaeographis.
A unique, novel lineage was detected clustering between Platythecium and Allographa. The new genus, Mangoldia, from Australia combines a Phaeographis morphology with Graphis type hymenium and ascospores, a previously unknown combination of characters (Lücking et al. 2012).
Subfamily Graphidoideae: Acanthothecis clade
This clade includes species of Acanthothecis s.str. with weakly corticate or ecorticate thallus and rounded ascomata, as well as some corticolous species currently classified in Topeliopsis: Topeliopsis darlingtonii and Topeliopsis elixii. The morphology of these taxa is quite similar and most likely they should all be included within Acanthothecis, although not all have the apically spinulose paraphyses and periphysoids characteristic of that genus (Staiger and Kalb 1999, Staiger 2002). Species of Acanthothecis s.lat. with corticate thallus and lirellate ascomata do not belong here but cluster next to Carbacanthographis.
Subfamily Graphidoideae: Acanthotrema clade
Acanthotrema is a small genus of four species characterized by a corticate thallus, chroodiscoid ascomata, and thin-walled ascospores of the Chroodiscus type (Rivas Plata et al. 2010b). This genus, which is found on shaded tree trunks in the lowland rain forest, appears rather isolated within the family on a very long branch and its exact position remains unresolved. Depending on taxon sampling, it either clusters close to tribe Thelotremateae or the Diploschistes clade (Papong et al. 2009, Rivas Plata et al. 2012a), but always lacking support for either placement.
Subfamily Graphidoideae: Carbacanthographis clade
This clade includes two strongly supported genus-level clades: Carbacanthographis and Acanthothecis 2, in addition to a further species of Carbacanthographis forming an unsupported sister clade. Carbacanthographis has graphidoid ascomata but is not closely related to Graphis and allies, whereas Acanthothecis 2 has fissurinoid-hemithecioid ascomata, while not being closely related to either Fissurina or Hemithecium (Staiger and Kalb 1999, Staiger 2002). Carbacanthographis currently includes close to 20 species, but only two have been sequenced; molecular data for more species are required to confirm monophyly of this genus. Only one species of Acanthothecis falling into this clade, Acanthothecis peplophora, has been sequenced so far, but its morphology suggests that this clade includes species of Acanthothecis with corticate thallus and striate lirellae, such as the widespread tropical Acanthothecis subclavulifera (Staiger and Kalb 1999, Staiger 2002). This clade will eventually require a new genus name; it is not closely related to Acanthothecis s. str., based on Acanthothecis hololeucoides.
Subfamily Graphidoideae: Diploschistes clade
Diploschistes is a medium-sized genus of about 30 currently accepted species (Lumbsch 1989, Guderley and Lumbsch 1996, Lumbsch and Elix 2003, Rivas Plata et al. 2010b). It is unique in the family in having a chlorococcoid photobiont and due to its ecology and chemistry, forming well-developed, thick thalli usually on soil or rock surfaces or sometimes on bryophytes with the depsides gyrophoric, lecanoric, and diploschistesic acids as main compounds. The genus usually clusters close to tribe Thelotremateae but this position is not supported. The very long stem branch together with the very short terminal branches suggest that this genus radiated very recently, which is remarkable considering that it is cosmopolitan in distribution. The placement of the norstictic acid containing, lecanoroid Diploschistes ocellatus requires additional studies (see below). Another genus supposedly related to Diploschistes, Ingvariella (Guderley et al. 1997), has been shown to belong in Stictidaceae (Fernández-Brime et al. 2011).
Subfamily Graphidoideae: Diploschistes ocellatus clade
Diploschistes ocellatus differs from other species of Diploschistes in the lecanoroid ascomata and norstictic acid as major compound in the thallus. Its phylogenetic position is unresolved, although it usually clusters close to or at the base of the Diploschistes clade (Fernández-Brime et al. 2011). Additional loci need to be sequenced to address monophyly of Diploschistes as currently circumscribed.
Subfamily Graphidoideae: Heiomasia clade
This clade includes a single genus with two known species, Heiomasia seaveyorum and Heiomasia sipmanii. It was recently described and both species are only known with isidioid propagules but lack ascomata (Nelsen et al. 2010). Additional loci need to be sequenced to address the relationships of this isolated clade but it might well represent a relict clade with no close extant relative, similar to the case of Phaeographopsis.
Subfamily Graphidoideae: Phaeographopsis clade
Phaeographopsis is a lowland rain forest genus. It resembles species of Diorygma in the green, ecorticate thallus and pruinose, lirellate ascomata, but differs in the thin-walled, dark brown ascospores which accumulate above the hymenium and form weakly developed mazaedia (Aptroot et al. 1997, 2007, Kalb 2004, Lücking and Rivas Plata 2008). The genus, which includes two species, consistently clusters close to Diploschistes but without support; therefore its phylogenetic position is unresolved. The unique morphological features and long branch suggest this to represent another relict taxon in Graphidaceae.
Subfamily Graphidoideae: Topeliopsis clade
This clade was previously included in tribe Thelotremateae (Rivas Plata et al. 2012a) but with this larger dataset falls outside that tribe. It includes the bulk of taxa with topeliopsidoid-schizotremoid ascomata, that is, apothecia with layered (striate) excipulum; many species occur in the southern Hemisphere or are tropical-montane and often grow on bryophytes (Frisch and Kalb 2006, Mangold et al. 2008b, Lumbsch et al. 2010). Within this clade, both Schizotrema s.str. (excipulum carbonized, ascomata erumpent over tree bark) and Topeliopsis s.str. (excipulum non-carbonized, ascomata mostly sessile on bryophytes) form monophyletic and well-supported clades. Melanotopelia (excipulum carbonized, ascomata mostly sessile on bryophytes) is also potentially monophyletic, but only one of the four currently accepted species has been sequenced so far. Two species of Schizotrema s.lat. from separate clades and their relationships need to be studied further. Two further species of Topeliopsis cluster within to Acanthothecis s. str.
Subfamily Graphidoideae: Wirthiotrema clade
This well-supported clade includes species previously classified in Thelotrema but apparently not related to the latter genus. In addition, the mazaediate Nadvornikia hawaiiensis is nested within this clade. Three supported subclades can be distinguished: a clade consisting of Thelotrema bicinctulum, a corticate species morphologically intermediate between Thelotrema and Myriotrema; a clade consisting of Chapsa platycarpa and Thelotrema leucophthalmum; a clade consisting of Myriotrema peninsulae, Leudodecton expallescens, and Nadvornikia hawaiiensis; and a well-supported clade representing the recently segregated genus Wirthiotrema (Rivas Plata et al. 2010a). This entire clade will be treated in detail in a separate, forthcoming paper.
Subfamily Graphidoideae: tribe Thelotremateae
This tribe is well-supported in all analyses and contains four currently accepted genera, including a monophyletic Chroodiscus forming sister to the remaining genera, a monophyletic Leucodecton, and a monophyletic Thelotrema s.str., all strongly supported. These three genera differ consistently in ascomata morphology: Chroodiscus has chroodiscoid ascomata lacking periphysoids, Leucodecton also lacks periphysoids but its ascomata are mostly myriotremoid-porinoid, and Thelotrema features mostly lepadinoid ascomata with periphysoids and double margin (proper excipulum free). On the other hand, the genus Chapsa, with chroodiscoid ascomata and periphysoids, is polyphyletic, forming several well-supported clades. There is some correlation with morphological features in these clades but these need to be studied further, and this clade is currently being analyzed in a separate study (Parnmen et al. 2012). Also, several species of Chapsa s.lat. fall outside this tribe, such as Chapsa platycarpa and Chapsa leprieurii. The well-supported placement of the latter as sister to Ocellularia s.lat. is another surprising find of this study.
Subfamily Graphidoideae: Leptotrema clade
This clade was previously included in tribe Ocellulariae as basal sister group to Ocellularieae s.str. (Rivas Plata and Lumbsch 2011, Rivas Plata et al. 2012a), but the larger dataset analysed here does not support this relationship, although the clade consistently comes out as sister to Ocellularieae. The clade includes two genera, Leptotrema and Reimnitzia, which agree in general thallus morphology and ascus and ascospore type (Frisch et al. 2006a), but differ in the myriotremoid versus chroodiscoid ascomata.
Subfamily Graphidoideae: tribe Ocellularieae
This tribe is being treated in detail in a parallel paper based on roughly the same dataset (Rivas Plata et al. 2012b) and therefore is not further discussed here. It includes well over 300 species currently assigned to up to 12 genera: Ampliotrema, Compositrema, Fibrillithecis, Glaucotrema, Gyrotrema, Melanotrema, Myriotrema, Ocellis, Ocellularia s.lat., Redingeria, Rhabdodiscus, and Stegobolus. However, several more lineages exist deserving generic status and some genera appear to be nested within others, such as Gyrotrema within Ocellularia s.str. We expect that a more complete dataset in terms of complete sampling of genes will help to resolve this clade. Very unexpected is the placement of Chapsa leprieurii in Ocellularia s.lat.; this species has Chapsa-like ascomata with lateral periphysoids, a character otherwise absent in the entire Ocellularieae. Yet, its placement is supported by several sequences representing all three genes from two different specimens. We have no explanation for this placement, other than recognizing it as yet another remarkable case of parallel evolution in Graphidaceae (Rivas Plata and Lumbsch 2011).
Species delimitations in Graphidaceae
Although the sampling for this study was focused in targeting as many species as possible, with generally few repeats per species, the data allows a first approach in testing species concepts. The identifications made for the purpose of this study already represent a refined species concept as result of cross-examination of all available material and comparison with authentic type specimens. Thus, for example, previous identifications made using a more coarse species concept had already been corrected prior to this analysis, particularly in the genera Thelotrema and Ocellularia s.lat. (e.g. Rivas Plata et al. 2012b). Still, of the 185 species represented by at least two (and up to 21) OTUs, 37 turned out to be paraphyletic or polyphyletic. Of these, 12 cases can be explained by insufficient gene sampling (non-overlapping genes for different OTUs of the same species). Another 13 cases can potentially be resolved by analysing the corresponding clades separately, since for terminal clades, gap-rich regions caused by aligning a multitude of sequenced across a large group of species will mask the phylogenetic signal contained in these regions. Thus, the remaining 12 out of 185 species appear to present problems in terms of species concepts. These may either represent cases of too narrow species concepts (e.g. Pallidogramme chlorocarpoides versus Pallidogramme chrysenteron, which differ only in ascospore size and number) or cases of cryptic species (e.g. Fissurina marginata, Ocellularia diacida, Thelotrema adjectum). In some cases, such as Glyphis cicatricosa and Graphis scripta, the sequenced material could not be completely tested, but available data suggest that more than one species are hidden under these names. For example, Glyphis cicatricosa includes forms with different ascomata configuration (rounded versus lirellate-stellate), which might explain the presence of more than one species. In the case of Graphis scripta, a recent revision (Neuwirth and Aptroot 2012) already divided this taxon into at least four species, and the molecular data seem to confirm this view. Other species, such as Fissurina aggregatula versus Fissurina pseudostromatica, or taxa such as Sarcographa ramificans and Thecaria montagnei, include specimens with subtle morphological differences and these are obviously not yet fully understood. Notable is the case of Topeliopsis acutispora versus Topeliopsis decorticans, two morphologically similar species that have quite different ascospores; the clade including these species obviously requires more scrutinity. In spite of these unresolved cases, it however seems that the combination of molecular analysis and critical revision of phenotype characters has improved the species concept in Graphidaceae considerably.
It was generally assumed that many lichens have a wide geographic distribution, much wider than vascular plants (Jørgensen 1983, Galloway 1988, Tibell 1994, Lücking 2003, Feuerer and Hawksworth 2007). However, molecular phylogenetic studies have challenged this view for many taxa (Tehler et al. 2009, 2010, Del-Prado et al. 2011, Molina et al. 2011), and Graphidaceae are no exception. While previous studies assumed a proportion of 30–50% intercontinental, pantropical, or even cosmopolitan species (Wirth and Hale 1963, 1978, Hale 1974, 1978, 1981), the present analysis only confirmed 31 out of 148 monophyletic species (21%) as belonging to one of these categories, and only 20 as pantropical. In part this is a sampling artifact, and with more sequence data available, more species, such as Leucodecton compunctellum and Myriotrema olivaceum (for which unsequenced material from the Neotropics is known), will be confirmed to have a wide distribution. However, this is probably more than counterbalanced by the large number of new species with apparently narrow distribution ranges discovered in recent, detailed inventories (Kalb 2009, Rivas Plata and Lücking 2012, Rivas Plata et al. 2012b, Sipman et al. 2012). It also appears that some of the pantropical species, such as Glaucotrema glaucophaenum, Platygramme caesiopruinosa, Thelotrema diplotrema, and Thelotrema pachysporum, exhibit a highly structured phylogeny, suggesting that these might represent species complexes with separate, (semi-)cryptic species occurring in different geographic regions. Therefore, it appears that the proportion of species with narrow distribution ranges in Graphidaceae, and in lichens in general, is much higher than previously assumed.