(C) 2012 H. Thorsten Lumbsch. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
The phylogenetic position of the Tasmanian endemic genus Cameronia Kantvilasis studied using partial sequences of nuclear LSU and mitochondrial SSU ribosomal DNA. Monophyly of the genus is supported, as is its placement in Ostropomycetidae, although its position within this subclass remains uncertain. Given the lack of close relatives to Cameronia and its morphological differences compared to other families with perithecioid ascomata in Ostropomycetidae, the new family Cameroniaceae Kantvilas & Lumbsch is proposed.
Cameroniaceae, lichens, new family, Tasmania, taxonomy
The lichen flora of Tasmania has a remarkable number of unique species, as well as several genera that are unknown or very rarely found in other regions. Examples include the genera Jarmania Kantvilas (
Lichen taxa unique to Tasmania include the genus Cameronia (
Thick-walled asci having a hemiamyloid wall and non-amyloid tholus, anastomosing paraphysoids and muriform ascospores are all characters reminiscent of Arthoniales (
The study is based on fresh material collected by GK and deposited in the Tasmanian Herbarium (HO) and the Field Museum of Natural History (F), and on DNA sequences downloaded from Genbank. Sequences of Umbilicariaceae were included as outgroup since this family has been shown previously to be sister to Lecanoromycetidae+Ostropomycetidae (
Sequences obtained from Genbank for the study. Family or generic group as in figure 1, largely following (
| Species | Family/generic group as in Fig. 1 | nuLSU | mtSSU |
|---|---|---|---|
| Acarosporina microspora | Stictidaceae | AY584643 | AY584612 |
| Agyrium rufum | - | EF81824 | EF81821 |
| Ainoa mooreana | - | AY212850 | AY212828 |
| Anzina carneonivea | - | AY212829 | AY212851 |
| Arctomia delicatula | Arctomiaceae | AY853307 | AY853355 |
| Arctomia teretiuscula | Arctomiaceae | DQ007346 | DQ007349 |
| Aspicilia caesiocinerea | Megasporaceae | DQ780303 | DQ780271 |
| Aspicilia cinerea | Megasporaceae | DQ780304 | DQ780272 |
| Aspicilia contorta | Megasporaceae | DQ986782 | DQ986876 |
| Aspicilia hispida | Megasporaceae | DQ780305 | DQ780273 |
| Baeomyces placophyllus | - | AY300878 | AF356658 |
| Baeomyces rufus | - | DQ871008 | DQ871016 |
| Belonia russula | Gyalectaceae | FJ941887 | AY648888 |
| Bryophagus gloeocapsa | Gyalectaceae | AF465440 | AY300880 |
| Cameronia pertusarioides 6504 | - | JX977114 | JX977110 |
| Cameronia pertusarioides 6505 | - | JX977115 | JX977111 |
| Cameronia pertusarioides 6506 | - | JX977116 | JX977112 |
| Cameronia tecta | - | JX977117 | JX977113 |
| Chapsa phlyctidioides | Graphidaceae | JX465300 | EU675275 |
| Chapsa pulchra | Graphidaceae | EU075619 | EU075571 |
| Coccomycetella richardsonii | Odontotremataceae | HM244761 | HM244737 |
| Coccotrema cucurbitula | Coccotremataceae | AF274092 | AF329161 |
| Coccotrema pocillarium | Coccotremataceae | AF274093 | AF329166 |
| Coenogonium leprieurii | Coenogoniaceae | AF465442 | AY584698 |
| Coenogonium luteum | Coenogoniaceae | AF279387 | AY584699 |
| Coenogonium pineti | Coenogoniaceae | AY300834 | AY300884 |
| Cryptodiscus pallidus | Stictidaceae | FJ904677 | FJ904701 |
| “Cryptodiscus” rhopaloides | - | FJ904685 | FJ904707 |
| Dibaeis baeomyces | Icmadophilaceae | AY789291 | AY584704 |
| Diploschistes cinereocaesius | Graphidaceae | AY300835 | AY300885 |
| Diploschistes scruposus | Graphidaceae | AF279389 | AY584692 |
| Dyplolabia afzelii | Graphidaceae | HQ639628 | HQ639594 |
| Elixia flexella | - | AY853368 | AY853320 |
| Fissurina insidiosa | Graphidaceae | DQ973045 | DQ972995 |
| Glyphis cicatricosa | Graphidaceae | HQ639630 | HQ639610 |
| Graphis scripta | Graphidaceae | AY853322 | AY853370 |
| Gregorella humida | Arctomiaceae | AY853329 | AY853378 |
| Gyalecta flotowii | Gyalectaceae | AY300838 | AY300889 |
| Gyalecta hypoleuca | Gyalectaceae | AF465453 | HQ659180 |
| Gyalecta truncigena | Gyalectaceae | HM244766 | HM244743 |
| Gyalecta ulmi | Gyalectaceae | AF465463 | AY300888 |
| Gyalectaria gyalectoides | Coccotremataceae | GU980983 | GU980975 |
| Gyalectaria jamesii | Coccotremataceae | GU980984 | GU980976 |
| “Gyalidea”praetermissa | - | HM244768 | HM244745 |
| Hymenelia lacustris | Hymeneliaceae | AY853371 | AY853323 |
| Icmadophila ericetorum | Icmadophilaceae | DQ883694 | DQ986897 |
| Lobothallia radiosa | Megasporaceae | DQ780306 | DQ780274 |
| Myriotrema olivaceum | Graphidaceae | EU075627 | EU075579 |
| Nadvornikia hawaiiensis | Graphidaceae | AY605080 | EU075581 |
| Ocellularia chiriquiensis | Graphidaceae | EU075629 | EU075582 |
| Ocellularia endoxantha | Graphidaceae | AY605082 | EU075589 |
| Ochrolechia androgyna | Ochrolechia | AY300846 | AY300897 |
| Ochrolechia balcanica | Ochrolechia | AF329171 | AF329170 |
| Ochrolechia frigida | Ochrolechia | AY300847 | AY300898 |
| Ochrolechia oregonensis | Ochrolechia | DQ780308 | DQ780276 |
| Ochrolechia pallescens | Ochrolechia | DQ780310 | DQ780277 |
| Ochrolechia parella | Ochrolechia | AF274097 | AF320173 |
| Ochrolechia peruensis | Ochrolechia | DQ780311 | DQ780279 |
| Ochrolechia turneri | Ochrolechia | AY568002 | AY567982 |
| Ochrolechia yasudae | Ochrolechia | DQ986776 | DQ986902 |
| Ochrolechia sp. | Ochrolechia | DQ986777 | DQ986886 |
| Odontotrema phacidiellum | Odontotremataceae | HM244769 | HM244748 |
| Odontotrema sp. | Odontotremataceae | HM244772 | HM244751 |
| Orceolina antarctica | Trapeliaceae | AY212852 | AF274115 |
| Orceolina kerguelensis | Trapeliaceae | AY212830 | AF381561 |
| Paschelkiella pini | Stictidaceae | HM244762 | HM244738 |
| “Pertusaria” albescens | Variolaria-group | AF329176 | AF329175 |
| “Pertusaria” amara | Variolaria-group | AF274101 | AY300900 |
| Pertusaria coccodes | Pertusariaceae | AF2741095 | AY567984 |
| “Pertusaria”corallina | Variolaria-group | AY300850 | AY300901 |
| “Pertusaria” corallophora | Variolaria-group | DQ780316 | DQ780285 |
| Pertusaria coronata | Pertusariaceae | AY300851 | AY300902 |
| Pertusaria gibberosa | Pertusariaceae | DQ780322 | DQ780289 |
| Pertusaria lecanina | Pertusariaceae | AF274296 | AY567991 |
| Pertusaria leioplaca | Pertusariaceae | AY300852 | AY300903 |
| “Pertusaria” mammosa | Variolaria-group | AY212831 | AY212854 |
| Pertusaria mesotropa | Pertusariaceae | DQ780325 | DQ780292 |
| “Pertusaria“ophthalmiza | Variolaria-group | AY568006 | AY567993 |
| Pertusaria paramerae | Pertusariaceae | DQ780326 | DQ780293 |
| Pertusaria pertusa | Pertusariaceae | AF279300 | AF381565 |
| Pertusaria plittiana | Pertusariaceae | DQ780328 | DQ780294 |
| Pertusaria pustulata | Pertusariaceae | DQ780332 | DQ780297 |
| “Pertusaria” scaberula | Variolaria-group | AF274099 | AF431959 |
| “Pertusaria” subventosa | Variolaria-group | AY300854 | AY300905 |
| Phlyctis agelaea | Phlyctidaceae | AY853381 | AY853332 |
| Phlyctis argena | Phlyctidaceae | DQ986771 | DQ986880 |
| Phyllobaeis erythrella | - | DQ986780 | DQ986888 |
| Placopsis cribellans | Trapeliaceae | DQ871010 | DQ871018 |
| Placopsis gelida | Trapeliaceae | AY212836 | AY212859 |
| Placopsis santessonii | Trapeliaceae | AY212845 | AY212867 |
| Placynthiella icmalea | Trapeliaceae | AY212846 | AY212870 |
| Placynthiella uliginosa | Trapeliaceae | DQ986774 | DQ986877 |
| Protothelenella corrosa | Protothelenellaceae | AY607734 | AY607746 |
| Protothelenella sphinctrinoidella | Protothelenellaceae | AY607735 | AY607747 |
| Pycnotrema pynoporellum | Graphidaceae | JX421615 | JX421295 |
| Rhexiophiale rhexoblephara | - | AY853391 | AY853341 |
| Schizoxylon albescens | Stictidaceae | DQ401144 | DQ401142 |
| Siphula ceratites | Icmadophilaceae | AY853394 | AY853344 |
| Schaereria corticola | - | AY300909 | AY300859 |
| Stegobolus subcavatus | Graphidaceae | EU075641 | EU075595 |
| Stictis populorum | Stictidaceae | AY527327 | AY300882 |
| Stictis radiata | Stictidaceae | AY300864 | AY584727 |
| Thamnolia vermicularis | Icmadophilaceae | AY853345 | AY853395 |
| Thecaria quassiicola | Graphidaceae | HQ639667 | JF828971 |
| Thelotrema lepadinum | Graphidaceae | AY300866 | AY300916 |
| Thelotrema subtile | Graphidaceae | DQ871013 | DQ871020 |
| Thelotrema suecicum | Graphidaceae | AY300867 | AY300917 |
| Topeliopsis decorticans | Graphidaceae | EU075654 | EU075609 |
| Trapelia chiodectonoides | Trapeliaceae | AY212847 | AY212873 |
| Trapelia placodioides | Trapeliaceae | AF274103 | AF431962 |
| Trapeliopsis flexuosa | Trapeliaceae | AF274118 | AY212875 |
| Trapeliopsis granulosa | Trapeliaceae | AF274119 | AF381561 |
| Trapeliopsis percrenata | Trapeliaceae | AF279302 | AY212876 |
| Umbilicaria crustulosa | Umbilicariaceae | AY300869 | AY300919 |
| Umbilicaria decussata | Umbilicariaceae | HM161603 | HM161628 |
| Umbilicaria hyperborea | Umbilicariaceae | AY853399 | AY853349 |
| Varicellaria hemisphaerica | Varicellaria | AF381563 | AF381556 |
| Varicellaria lactea | Varicellaria | AF381557 | AF381564 |
| Varicellaria velata | Varicellaria | AY300855 | AY300906 |
| Wawea fruticulosa | Arctomiaceae | DQ007347 | DQ871023 |
We assembled partial sequences using Geneious Pro 5.4.3 (
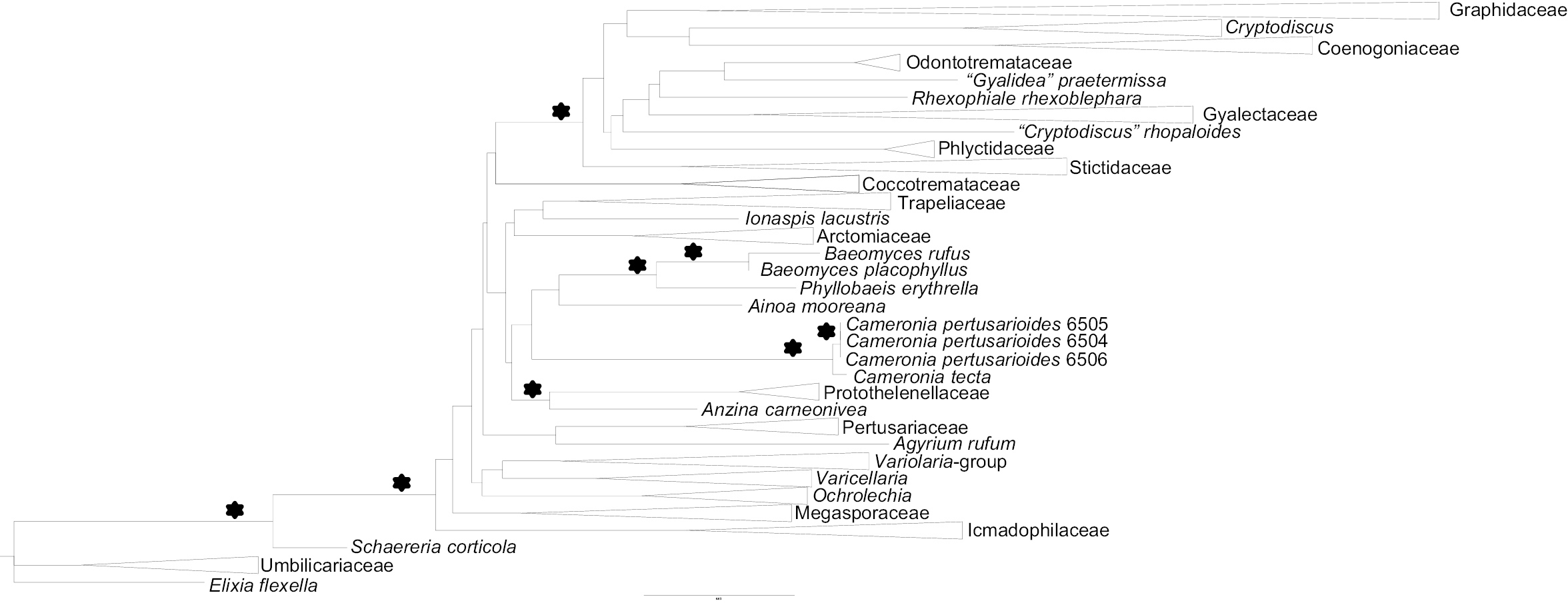
Eight new sequences were generated for this study and aligned with sequences downloaded from Genbank (Table 1). The single gene locus trees did not show any conflicts and hence the concatenated data set was analyzed. Our combined data set included 1313 unambiguously aligned positions, 370 of which were constant. The ML tree had a likelihood value of –26318.540 and in the B/MCMC analysis of the combined data set, the likelihood parameters in the sample had the following mean (Variance): LnL = -27045.138 (0.35). The ML tree and the tree from the B/MCMC tree sampling were almost identical, with no differences in well-supported clades. Furthermore, taxon sampling was very similar to that of previous studies focusing on the phylogeny of Ostropomycetidae (
Phylogenetic placement of Cameronia as inferred from a concatenated alignment of mtSSU and nuLSU DNA sequences. This is a simplified cartoon of the optimal tree under maximum likelihood with well supported families and species groups collapsed that were shown in previous studies (
The backbone of the Ostropomycetidae tree largely lacks support and the relationships of Cameronia within Ostropomycetidae remain unclear. Cameronia is the sister-group of Baeomycetaceae (Ainoa, Baeomyces, Phyllobaeis) but this relationship lacks support. This clade forms a sister-group to a well-supported clade that includes Anzina and Protothelenellaceae, but again, this relationship lacks support.
Although the molecular data support the placement of Cameronia in Ostropomycetidae, they fail to identify any close relatives of the genus, which is also reflected in the similarities of Blast searches of the newly generated sequences (maximal identity - nuLSU: 94%, mtSSU: 93%). Cameronia is distinguished by several characters that are generally used to characterize families, as shown in Table 2 where salient features of Cameronia and other families of Ostropomycetidae with perithecioid ascomata (Porinaceae, Protothelenellaceae, Thelenellaceae) are compared. The ascus type is very different from any of the other perithecioid Ostropomycetidae and also different from the apotheciate Baeomycetaceae, which have cylindrical asci (
Diagnostic features of families with perithecioid ascomata in Ostropomycetidae (
| Characters | Cameronia | Porinaceae | Protothelenellaceae | Thelenellaceae |
|---|---|---|---|---|
| Proper exciple | rudimentary | Well developed, consisting of periplectenchymatous cells | Well developed, consisting of periplectenchymatous to isodiametric cells | Well developed, consisting of periplectenchymatous cells |
| Hamathecium | Richly branched, anastomosing paraphysoids, no periphyses | Simple to sparsely branched Paraphyses, no periphyses | Richly branched, anastomosing paraphysoids, no periphyses | Richly branched, anastomosing paraphysoids, periphyses present |
| Asci | Broadly obovate | cylindrical | cylindrical | cylindrical |
| Tholus | Well-developed | Poorly developed | Well-developed | Poorly developed |
| Ascus amyloidity | Outer wall hemiamyloid, tholus non-amyloid | Non-amyloid | Outer and wall and tholus amyloid | Non-amyloid |
| Ocular chamber | - | - | + | +/- |
| Ascospores | Hyaline, non-halonate, thick-walled, muriform | Hyaline, halonate, thin- to thick-walled, transversely septate to muriform | Hyaline, halonate, thick-walled muriform | Hyaline to brownish, halonate, thin-walled, muriform |
| Chemistry | Dibenzofuranes, triphenyl | Nil or pigments | nil | nil |
Given the dissimilarity in morphological characters and the lack of close relatives in the phylogenetic tree, we propose a new family Cameroniaceae below to accommodate the genus Cameronia. The new family is placed in Ostropomycetidae with unclear ordinal position.
Mycobank no. MB802404
Cameronia Kantvilas, Lichenologist 44: 92. 2012.
Thallus crustose, photobiont a coccoid green alga. Ascomata perithecioid, immersed in the thallus, proper exciple rudimentary, hamathecium consisting of richly branched, anastomosing paraphysoids, inspersed with oil droplets, containing hymenial algae, periphyses absent. Asci broadly obovate, with outer wall hemiamyloid and with a well-developed, non-amyloid tholus; ocular chamber lacking. Ascospores hyaline, non-halonate, eumuriform. Conidiomata immersed in the thallus, forming baciliform to bone-shaped conidia.
This study was supported by the NSF grant “ATM – Assembling a taxonomic monograph: The lichen family Graphidaceae” (DEB-1025861). The laboratory work was done at the Pritzker Laboratory for Molecular Systematics at the Field Museum. For companionship in the field in quest of fresh material for analysis, GK thanks Brigitte de Villiers.