(C) 2012 Nicolas Magain. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Arctomia borbonica sp. nov. is described as new for science from montane natural and secondary habitats in Reunion in the Mascarene archipelago (Indian Ocean). It has a sterile, foliose, usually wrinkled, thallus whose margins produce goniocysts that disintegrate into a soredioid margin; it looks like a Leptogium species. Its phylogenetic position in the Arctomiaceae (Ostropomycetidae, Ascomycota) has been determined with 3 genes (nuLSU, mtSSU, RPB1) inferences.
Ascomycota, Ostropomycetidae, Arctomiaceae, Arctomia, phylogenetic inferences, nuLSU, mtSSU, RPB1, Reunion, Mascarene archipelago
Within the Lecanoromycetes, the subclass Ostropomycetidae Reeb, Lutzoni and Cl. Roux exhibits an impressive diversity of ascomata, thallus forms and ecological requirements. The phylogenetic relationships between genera and families are poorly resolved (
Within such a large and very much unresolved variation, the case of the Arctomiaceae is rather simple. The family is strongly supported and includes three genera: Gregorella and Wawea, each with one species, and Arctomia with five species (
We here report the discovery of a further new species, which we assign to the genus Arctomia, found epiphytic in montane habitats in the island of Reunion (Mascarene archipelago, Indian Ocean). The material was first assigned to Leptogium, a genus belonging to the Collemataceae in the Lecanoromycetidae (
Well-preserved lichen specimens lacking any visible symptoms of fungal infection were used for DNA isolation. Extraction of DNA and PCR amplification were performed following the protocol of
We assembled matrices with most representatives of species included by
Species and specimens used for this study, with GenBank accessions numbers for the three loci examined. Newly produced sequences for Arctomia borbonica are in bold.
| Species name | LSU | mtSSU | 1RPB |
|---|---|---|---|
| Absconditella sp. | AY300825 | AY300873 | — |
| Acarosporina microspora | AY584643 | AY584612 | DQ782818 |
| Agyrium rufum | EF581826 | EF581823 | EF581822 |
| Arctomia borbonica 1 (holotype) | JX030030 | JX030032 | JX030034 |
| Arctomia borbonica 2 | JX030031 | JX030033 | JX030035 |
| Arctomia delicatula | AY853355 | AY853307 | DQ870929 |
| Arctomia interfixa | DQ007345 | DQ007348 | — |
| Arctomia teretiuscula | DQ007346 | DQ007349 | DQ870930 |
| Aspicilia contorta | DQ986782 | DQ986876 | DQ986852 |
| Bacidia rosella | AY300829 | AY300877 | AY756412 |
| Chromatochlamys muscorum | AY607731 | AY607743 | FJ941910 |
| Coccotrema pocillarium | AF274093 | AF329166 | DQ870940 |
| Conotrema populorum | AY300833 | AY300882 | — |
| Diploschistes ocellatus | HQ659183 | HQ659172 | DQ366252 |
| Gregorella humida | AY853378 | AY853329 | — |
| Gyalectaria diluta | GU980982 | GU980974 | — |
| Icmadophila ericetorum | DQ883694 | DQ986897 | DQ883723 |
| Lecanora intumescens | AY300841 | AY300892 | AY756386 |
| Neobelonia sp. | AY300830 | AY300879 | — |
| Ochrolechia parella | AF274097 | AF329173 | DQ870959 |
| Ochrolechia upsaliensis | GU980986 | GU980979 | GU981009 |
| Orceolina kerguelensis | AY212830 | AY212853 | DQ870963 |
| Pertusaria amara | AF274101 | AY300900 | DQ973048 |
| Pertusaria lactea | AF381557 | AF381564 | DQ870971 |
| Pertusaria leioplaca | AY300852 | AY300903 | DQ870973 |
| Pertusaria paramerae | DQ780326 | DQ780293 | GU981012 |
| Pertusaria pertusa | AF279300 | AF381565 | DQ870978 |
| Pertusaria pustulata | DQ780332 | DQ780297 | GU981013 |
| Pertusaria subventosa | AY300854 | DQ780302 | DQ870981 |
| Placopsis gelida | AY212836 | AY212859 | DQ870984 |
| Protothelenella corrosa | AY607734 | AY607746 | DQ870988 |
| Protothelenella sphinctrinoidella | AY607735 | AY607747 | DQ870989 |
| Thamnolia vermicularis | AY853395 | AY853345 | DQ915599 |
| Thelotrema subtile | DQ871013 | DQ871020 | DQ870998 |
| Toninia cinereovirens | AY756365 | AY567724 | AY756429 |
| Trapelia chiodectonoides | AY212847 | AY212873 | DQ870999 |
| Trapeliopsis granulosa | AF274119 | AF381567 | DQ871001 |
| Wawea fruticulosa | DQ007347 | DQ871023 | DQ871005 |
Three matrices were assembled: 38 species with 927 included characters for nuLSU, 38 species with 668 included characters for mtSSU and 32 species with 675 included characters for RPB1 (part 1). Incongruence between the matrices was tested with maximum likelihood analysis using GARLI (
An unweighted maximum parsimony (MP) analysis was performed in PAUP* 4.0b10 (
A partition of six subsets was implemented in the concatenated matrix: nuLSU, mtSSU, intron in RPB1, and three for each RPB1 codon position. Models of evolution for the maximum likelihood and Bayesian analysis were selected based on the Akaike Information Criterion (
We tested the monophyly of the genus Arctomia by comparing the best unconstrained tree with the best tree obtained by constraining all Arctomia sequences to form a monophyletic group. Trees were generated in RaxML and then tested with two methods: the Shimodaira-Hasegawa (SH) test and the Expected Likelihood Weight (ELW) test as implemented in Tree-PUZZLE 5.2. (
The concatenated matrix with aligned sequences for nuLSU, mtSSU and RPB1 has 2781 characters, out of which 511 are excluded (330 for nuLSU out of which 250 represent introns in Bacidia rosella, 173 for mtSSU and 8 for RPB1), 983 are constant, 276 are parsimony-uninformative and 1011 are parsimony potentially informative. The most parsimonious tree has the following characteristics: length = 6295 steps, CI = 0.336 and RI = 0.428. The ML analysis yielded a tree with a likelihood value of Ln = -28660.4 and length of 6.175. Parameters of the partitions were as follows: LSU — p(A)= 0.2604, p(C)= 0.2216, p(G)= 0.2980, p(T)= 0.2199 a= 0.3134, r(A-C)= 0.7438, r(A-G)= 1.8229, r(A-T)= 0.7430, r(C-G)= 0.7409, r(C-T)= 4.5270, r(G-T)= 1.0000; mtSSU — p(A)= 0.3330, p(C)= 0.1606, p(G)= 0.2136, p(T)= 0.2926, a= 0.4207, r(A-C)= 0.9284, r(A-G)= 2.9298, r(A-T)= 1.6160, r(C-G)= 0.6649, r(C-T)= 3.4571, r(G-T)= 1.0000; RPB1 intron — p(A)= 0.2349, p(C)= 0.2056, p(G)= 0.2267, p(T)= 0, 3287, a= 0.9412, r(A-C)= 6.9358, r(A-G)= 21.9085, r(A-T)= 11.1853, r(C-G)= 8.6280, r(C-T)= 19.3378, r(G-T)= 1.0000; RPB1, 1st codon — p(A)= 0.2778, p(C)= 0.2440, p(G)= 0.3318, p(T)= 0.1463, a= 0.4211; r(A-C)= 4.0125, r(A-G)= 5.8268, r(A-T)= 3.1946, r(C-G)= 2.7176, r(C-T)= 2907386, r(G-T)= 1.0000; RPB1, 2nd codon — p(A)= 0.3521, p(C)= 0.2038, p(G)= 0.2319, p(T)= 0.2122, a= 0.3474, r(A-C)= 1.7253, r(A-G)= 3.1209, r(A-T)= 0.5159, r(C-G)= 1.9509, r(C-T)= 4.4498, r(G-T)= 1.0000; RPB1, 3rd codon — p(A)= 0.2683, p(C)= 0.2056, p(G)= 0.2545, p(T)= 0.2716, a= 0.5667, r(A-C)= 8.7546, r(A-G)= 24.9090, r(A-T)= 4.6296, r(C-G)= 5.8128, r(C-T)= 56.3087, r(G-T)= 1.0000.
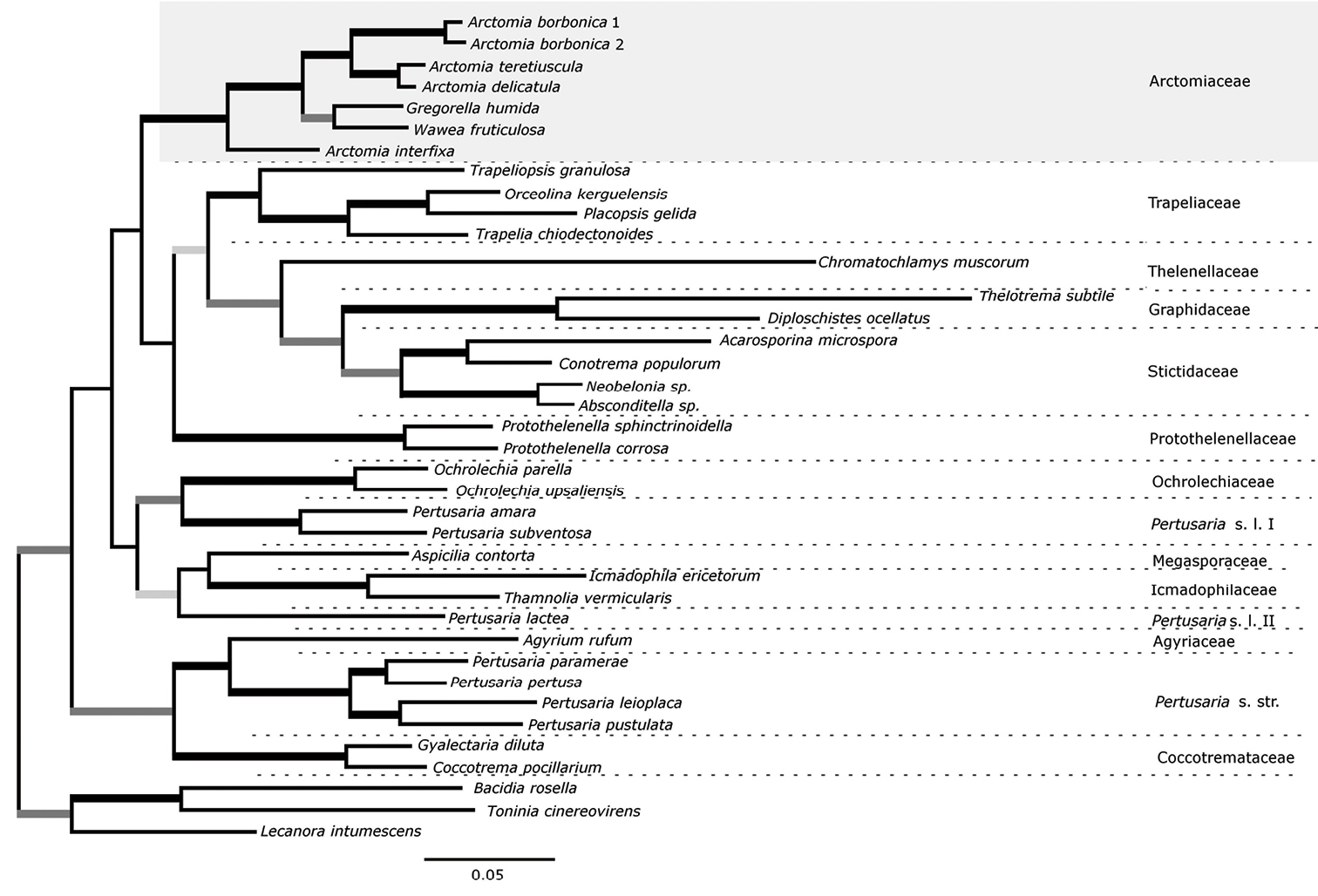
All three analyses retrieve the family Arctomiaceae as a strongly supported clade (MPBS= 81%, MLBS = 97%, PP=1) (Fig. 1). All nodes within the Arctomiaceae clade are strongly supported: Arctomia delicatula and Arctomia teretiuscula form a clade supported with MLBS= 99% and PP=1.0; they further form a clade with both accessions of Arctomia borbonica that is supported with MLBS = 94% and PP=1.0; Gregorella humida and Wawea fruticulosa form a clade supported with MLBS = 86% and PP= 1.0; and finally the latter is sister to the clade including all accessions of Arctomia (except for Arctomia interfixa) in a node supported by MLBS= 95% and PP= 1.0.
50% consensus tree produced by the Bayesian analysis of a concatenated matrix with three loci (nuLSU, mtSSU and RPB1) with 2531 characters and highlighting the Arctomiaceae and the newly described Arctomia borbonica. Branches supported by MPBS and MLBS > 70% and Bayesian posterior probabilities > 0.95 are in black; those supported by MLBS >70% and Bayesian posterior probabilities > 0.95 in dark grey and those only by Bayesian posterior probabilities > 0.95 in light grey.
SH test shows that the likelihood of the topology constraining all Arctomia sequences to form a monophyletic group is not significantly worse (at 0.05 significance level) than that with Arctomia interfixa being sister to all other accessions of the Arctomiaceae. Following that test, the monophyly of all species assigned to Arctomia, incl. Arctomia borbonica sp. nov., cannot be rejected. The result of the ELW is the contrary: such a monophyly is rejected at 0.0473 significance level.
DiscussionThe lichen family Arctomiaceae is fully recovered in our analysis (Fig. 1) and all other accessions are resolved in positions fully consistent with those published for the Ostropomycetidae (
Diagnostic characters for the genera recognized within the Arctomiaceae are given by
The hypothesis of describing a new genus for Arctomia borbonica has been carefully assessed. Indeed, the genus as circumscribed by
Species recognized by its foliose, usually much crumpled, blue grey to brown thallus producing goniocysts at its margins, eventually forming a soredioid margin. Ascomata and conidiomata unknown.
REUNION (Mascarene archipelago). Forêt de Bébour, track starting at Gîte de Bélouve toward Piton des Neiges, 21°4'49"S, 55°31'24"E (DMS), 1850 m alt., 9 Nov 2009, wet montane ericoid tickets, N. Magain & E. Sérusiaux sn (holotype : LG).
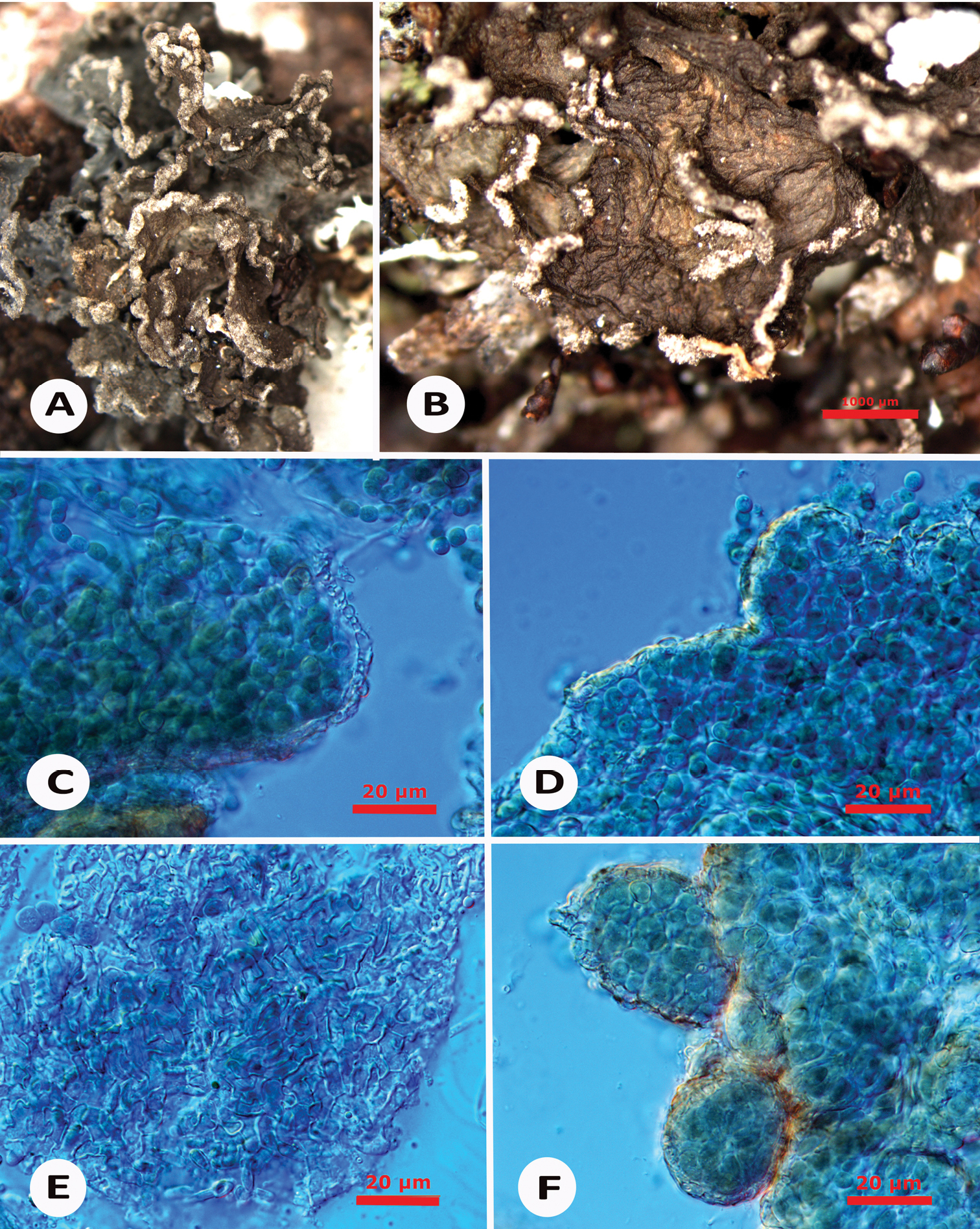
Thallus not exceeding 1 cm in diam., with distinct lobes when well-developed, lobes blue-grey to brown when dry, up to 0.2-0.3 mm wide and c. 200-400 µm thick, hardly distinguished in some specimens, with a surface typically wrinkled (even in young lobes), sometimes very much “crumpled”, always developing small goniocysts, mainly at the margins but also on the upper surface; cortex (Fig. 2C–E) developed on upper and lower sides, formed by a single layer of small rounded (in cross section) and jigsaw-like (in surface view) cells, less than 5 µm thick; goniocysts (Fig. 2F) 20-80 µm across, always containing compact chains of Nostoc cells and covered by a layer of isodiametric to rounded cells, 2–5 µm, best developed at the lobes margins where they eventually form a typical pale brownish soredioid edge, due to cortical disintegration. Photobiont belonging to the cyanobacteria genus Nostoc forming chains of small rounded cells 2–5 µm in diam. Ascomata and conidiomata unknown.
Arctomia borbonica (holotype). A–B macroscopic view of the thallus, with details of the wrinkled surface B and soredioid margin, made of disintegrating goniocysts C–D cross section through the thallus, showing the cortex with small, isodiametric cells, and the Nostoc chains E surface view of the cortex F young goniocysts formed at the lobes margins. Scale: A–B = 1 mm; C–E = 20 µm.
No secondary metabolites found by TLC.
The material looks like a species in Leptogium, a genus belonging to the Collemataceae in the Lecanoromycetidae (
Arctomia borbonica has been collected at three different sites on the island of Reunion in the Mascarene archipepago, incl. in highly disturbed secondary tickets with Eucalyptus plantations; it grows on trunks (Eucalyptus, Acacia heterophylla) or on main stems of Erica tickets. It is probably widespread on the island. The two localities with natural vegetation belong to two different and typical habitats. The first one is the margin of the “Forêt de tamarins des hauts” with large boles of the endemic tree Acacia heterophylla (locality at the nature reserve “Roche Ecrite”, at 1500 m) and corresponds to the “Acacia mountain forest” in
REUNION (Masarenes archipelago). Nature reserve at Roche Ecrite, track to the summit, 20°58'6"S, 55°26'26"E (DMS), c. 1500 m alt., 4 nov 2009, montane forest dominated by Acacia heterophylla, N. Magain & E. Sérusiaux sn (LG). S part of the island, N of St-Philippe, near « gîte Bernard Brice », 21°20'23"S, 55°41'55"E (DMS), 650 m alt., 10 Nov 2009, Eucalyptus plantations and secondary tickets, N. Magain & E. Sérusiaux sn (LG).
Field studies in Reunion were made possible with the help and advice from the Parc National de La Réunion, especially through the courtesy of Mr B. Lequette and Mr J. M. Pausé. Dr Cl. Ah-Peng and Prof. D. Strasberg of the University of La Réunion in Saint-Denis and Dr. J. Hivert of the Conservatoire Botanique National de Mascarin (St-Leu) were also very helpful. We thank them all most sincerely. We further thank Mr I. Cremasco and L. Gohy for technical assistance in the molecular laboratory and herbarium at the University of Liège. Nicolas Magain is a Ph.D. Student at the University of Liège and acknowledges the financial support by FRIA, an organ of the Belgian Research Foundation. Finally we thank both referees for their critical and helpful notes and suggestions.