(C) 2012 Paul F. Cannon. This is an open access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC-BY), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
For reference, use of the paginated PDF or printed version of this article is recommended.
Volutella colletotrichoides is shown to belong to the Plectosphaerellaceae rather than the Hypocreales where other species of that genus reside. The new genus Lectera is described for Volutella colletotrichoides, and for a further, previously undescribed species with slightly longer conidia and differences in rDNA ITS and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) sequences. Lectera species are found to be soil-dwelling organisms associated particularly with species of Fabaceae, and Lectera colletotrichoides is demonstrated to be a pathogen of Cicer arietinum and other grain legumes.
Systematics, plant pathogenic fungi, phylogeny, Cicer arietinum
Volutella colletotrichoides was first described from diseased alfalfa and other forage legumes collected from Iowa, USA (
The pathogenicity of Volutella colletotrichoides was demonstrated by observations of naturally infected plants in the glasshouse and in the field, and also by infection experiments (
Host range experiments (
Published reports of Volutella colletotrichoides from other regions are sparse. It was reported from Glycine max in Ethiopia by
In the winter of 1988, a field survey was carried out for diseases of chickpea (Cicer arietinum) in the vicinity of Salheia, at that time a newly reclaimed region between the Nile Delta and the Suez Canal (Ismaelia) in northern Egypt. There, blighted chickpea plants were observed and collected, with the causal organism initially assumed to be the common chickpea pathogen Ascochyta rabiei. The macroscopic visible symptoms were circular or elongated dark brown to black anthracnose-like lesions approximately one centimeter long on the lower stem parts in addition to partly chlorotic leaves. The causal organism was isolated and sent to the Danish Government Institute of Seed Pathology for Developing Countries (DGISP), and designated as DGISP 271. Later the diagnostic studies were transferred to The Danish Veterinary and Agricultural University. Here the organism was registered as CP 2035. After an extended period of analysis using both morphological and molecular methods with multiple collaborators, the affinities of CP 2035 have finally been confirmed to be with Volutella colletotrichoides.
As
Isolates were retrieved from storage in the CABI Genetic Resources Collection (see Table 1). For morphological analysis, strains were grown at 25°C for 7 d on PCA and PDA media (
Newly sequenced strains in this study
| Reference no. | Species | Host/substratum, geographical origin | ITS | GAPDH |
|---|---|---|---|---|
| IMI 185380 | Acremonium nepalense | Pinus sp. soil, Nepal | JQ647430 | |
| IMI 337226 | Bionectria coronata | Serjania sp., Venezuela | JQ700569 | |
| IMI 46339 | Gibellulopsis nigrescens | Soil, UK | JQ647441 | JQ724142 |
| IMI 118380 | Gibellulopsis nigrescens | Beta vulgaris, Canada | JQ647440 | JQ724134 |
| IMI 265740 | Lectera colletotrichoides | Soil, Brazil | JQ647449 | JQ724136 |
| IMI 303685 | Lectera colletotrichoides | Capsicum annuum, Morocco | JQ647450 | JQ724140 |
| IMI 332702 | Lectera colletotrichoides | Cicer arietinum, Egypt | JQ647428 | |
| IMI 366179 | Lectera colletotrichoides | Phaseolus vulgaris, Ethiopia | JQ693168 | |
| IMI 368065 | Lectera colletotrichoides | Xanthium spinosum, Argentina | JQ647451 | JQ724141 |
| IMI 181698 | Lectera longa | Triticum sp., Australia | JQ647448 | JQ724135 |
| IMI 312228 | Plectosphaerella cucumerina | Daucus carota, Denmark | JQ647434 | JQ724139 |
| IMI 49467b | Pseudonectria buxi | Buxus sempervirens, UK | JQ693165 | |
| IMI 61338 | Pseudonectria buxi | Buxus sp., UK | JQ647445 | |
| IMI 311153 | Pseudonectria buxi | Buxus sempervirens, France | JQ693164 | |
| IMI 322095 | Verticillium dahliae | Alectryon conaceum, UK | JQ647437 | JQ724138 |
| IMI 364520 | Verticillium dahliae | Prunus persica, Yugoslavia | JQ647438 | JQ724137 |
| IMI 62131 | Verticillium nonalfalfae | Lycopersicon esculentum, UK | JQ647435 | JQ724134 |
| IMI 67913 | Volutella ciliata | Pisum sativum, UK | JQ647447 | |
| IMI 74420 | Volutella ciliata | unknown host, Germany | JQ693167 | |
| IMI 341382 | Volutella ciliata | Sclerotinia sclerotiorum, locality unknown | JQ693166 | |
| IMI 341383 | Volutella ciliata | Sclerotinia sclerotiorum, locality unknown | JQ647446 | |
| IMI 332191 | Volutella consors | unknown source, India | JQ693163 | |
| IMI 92688 | Volutella lini | unknown host, India | JQ647452 | |
| IMI 224502 | Volutella lini | unknown host, India | JQ693169 | |
| IMI 136704 | Volutella ramkumarii | Submerged petioles, Papua New Guinea | JQ647453 |
Infection studies were carried out, using chickpea cultivar Family 88 as well as certain other species of Fabaceae as hosts: Vicia faba, Glycine max, Pisum sativum, Vigna unguiculata, and Phaseolus vulgaris. The inoculum was produced as above. Stems and leaves were inoculated without prior wounding using a spore suspension produced by flooding four week old cultures with 10 ml sterile distilled water. The conidial concentration was adjusted to 2 × 105 ml-1 and finally one drop of Tween 20 (as detergent) was added. Symptoms were observed two weeks after inoculation.
For the molecular analysis, strains were grown on malt extract (MADW) agar plates (
Total genomic DNA was obtained from a small loopful (1 µl) of each strain using a proprietary complex DNA release solution (microLYSIS®–PLUS; Microzone Ltd, UK) in accordance with manufacturer’s instructions. The thermal cycler lysis profile was: 15 min at 65°C, 2 min at 96°C, 4 min at 65°C, 1 min at 96°C, 1 min at 65°C, 30 s at 96°C and hold at 20°C.
Partial ribosomal RNA gene clusters (part of 18S small subunit RNA gene, internal transcribed spacer 1 (ITS1), 5.8S ribosomal RNA gene, internal transcribed spacer 2 (ITS2), part of 28S large subunit ribosomal RNA gene) were amplified by polymerase chain reaction (PCR) using primer set TW81 (fwd): 5’–GTTTCCGTAGGTGAACCTGC–3’ & AB28 (rev): 5`–ATATGCTTAAGTTCAGCGGGT–3’ (
Aliquots (4 µl) of amplification products were assessed for quality by gel electrophoresis using 1.5 % Seakem LE agarose (BMA, UK) for 2 h at 5V cm-1 in half-strength Tris-Borate-EDTA buffer (i.e., 0.5 × TBE buffer; 45 mM Tris; 45 mM Boric acid; 1.25mM EDTA, pH 7.5, see
Remaining unused PCR products were purified with the microCLEAN PCR Purification Kit (Microzone Ltd, UK) following the manufacturer’s instructions. The purified PCR products were utilised in sequencing reactions undertaken in a Primus 96 plus thermal cycler (MWG-BIOTECH AG, Germany) by using BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) with primer TW81 (as above). Sequencing conditions were: 96°C for 1 min followed by 25 cycles of 20 s at 96°C, 10 s at 50°C, 4 min at 60°C (ramp rate: 1°C s-1). Excess unincorporated BigDye was removed with DyeEx 2.0 affinity columns (Qiagen Ltd., UK) according to the manufacturer’s instructions and the sequencing reaction products were suspended in HiDi Formamide (Applied Biosystems, UK). These products were separated on a capillary array 3130 Genetic Analyser (Applied Biosystems, UK).
Sequence trace files were first assessed for quality using Sequencing Analysis Software v5.2 Patch 2 (Applied Biosystems, UK) and then checked manually using the software package Chromas v2.23 (Technelysium, Australia) and exported as text files. Sequences were deposited in GenBank and Accession numbers obtained (see Table 1).
Sequences of studied species were aligned using the default parameters of Clustal W (
Besides Cicer arietinum (from which the tester strain CP2035 (IMI 332702) was isolated), the following leguminous plants were found to function as hosts after experimental inoculation: Glycine max, Phaseolus vulgaris, Pisum sativum, Vicia faba and Vigna unguiculata. The main symptoms were evident as brown to black lesions on the lower stem parts, within which acervuli develop. Leaves also showed similar symptoms in all species, in addition to chlorosis. The symptoms were very similar to those caused by Ascochyta rabiei, a well-known and commonly occurring pathogen causing ascochyta blight on chickpea.
TaxonomyNamed after Dr Hannibal Lecter (
Conidiomata intermediate between sporodochia and acervuli, erumpent through host tissuesand without a clear upper wall, hemispherical to ± globose, pink or flesh-coloured, accompanied by dark brown septate tapering, primarily marginal, setae. Conidia formed from hardly modified conidiogenous cells, hyaline, aseptate, slightly curved, smooth, fusiform with pointed ends. Appressoria formed after conidial germination, dark brown, round to ovate with smooth margins.
Lectera colletotrichoides (Chilton) P.F. Cannon
Mycobank: MB 550042
Conidiomata formed at the apex of a short peg-like vegetative structure that is erumpent through host tissues, 80–350 µm diam, cushion-shaped to almost globose, with a compact palisade of conidiogenous cells usually surrounded by setae. Setae variably produced, with some conidiomata dark grey in coloration due to copious setae while others are pale pink or flesh-coloured with few or no setae; 50–130 µm in length, 3–6 µm in diam at the base, gradually tapering, golden to dark brown, smooth or sparsely verrucose, 2- to 3-septate, the apex acute. Conidiogenous cells 15–32 × 3–5 µm, cylindrical or slightly tapering with the apex rounded, proliferating percurrently with inconspicuous periclinal thickening and sometimes a minute collarette. Conidia inoculated onto Medicago sativa stem (6.5-) 7–10 (-11.5) [mean 8.35 µm, sd 0.73, n = 140] × 2.5–3 (-3.5) µm [mean 2.67 µm, sd 0.24, n = 140], mean length/width ratio 3.14: 1, cylindrical to cylindric-fusiform or navicular, the ends acute, slightly usually inaequilateral with one longitudinal face ± flat, hyaline, aseptate, smooth-walled, without a gelatinous sheath or appendages. Cultures on PCA and PDA at 25°C under alternating daylight/near UV growing moderately slowly, reaching 25–30 mm after 7 d, bright orange with a waxy appearance, the central part becoming brownish after 14 d, aerial mycelium poorly developed. Conidiomata absent or poorly developed, with setae fewer, narrower, shorter and less pigmented than in colonies on host tissue, and conidiogenous cells often formed singly at the apex of vegetative hyphae. Conidia 6.5–9 (-10.5) [mean 7.41 µm, sd 0.68, n = 100] × 2–3 µm [mean 2.43 µm, sd 0.22, n = 100], mean length/width ratio 3.07: 1, similar in appearance to those formed after inoculation on Medicago sativa stems. Appressoria 4.5–8 × 4–6.5 µm, circular to ovate with entire margins, dark brown. Sclerotia not observed.
USA: Iowa, Ames, on stems of Medicago sativa, Oct. 1954, J. Chilton(ISC 217496, lectotype, here designated; K(M) 176269, isolectotype of Volutella colletotrichoides). USA: Iowa, Ames, on stems of Medicago sativa, Oct. 1954, J. Chilton (ISC 217482, lectotype, here designated; K(M) 176270, isolectotype of Volutella colletotrichoides var. setosa). Other authentic material: same collection data, ISC 217488, ISC 217497 and ISC 217498 – syntypes of Volutella colletotrichoides; same collection data, ISC 217483 and ISC 217484 – syntypes of Volutella colletotrichoides var. setosa.
Associated with Asteraceae (Xanthium spinosum), Fabaceae (Cicer arietinum, Glycine max, Lotus corniculatus [Chilton, 1954], Medicago falcata [Chilton, 1954], Medicago sativa, Phaseolus vulgaris, Pisum sativum, Senna sophera, Trifolium hybridum [Chilton, 1954], Trifolium pratense [Chilton, 1954], Trifolium subterraneum, Vicia faba and Vigna unguiculata), Lamiaceae (Tectona grandis), Poaceae (Agrostis stolonifera, Hordeum vulgare, Triticum sp. and Zea mays), Solanaceae (Capsicum annuum) and Violaceae (Viola sp.).
Africa (Democratic Republic of the Congo, Egypt, Ethiopia, Morocco, Nigeria, South Africa [http://www.cbs.knaw.nl/], Zimbabwe [unpublished IMI record without voucher material]). Asia (India, Kuwait [unpublished IMI record without voucher material], Turkey [
Strains of Lectera colletotrichoides have been demonstrated to be pathogenic towards a range of Fabaceae species, but they are also commonly found associated with plants from other families and isolated from soil and plant litter. It also grows well in standard agar culture. It therefore can be presumed to exist (and probably grow actively) as a saprobe, and it is possible that the non-legume isolates originate from soils used to grow legumes in rotation.
The species as currently circumscribed has not been reported since 2002 but is very widely distributed, is associated with a wide range of plant taxa and apparently can exist as a saprobe in soil and leaf litter without a direct plant association. It does not appear to be an economically important pathogen except perhaps in limited circumstances. However, as there is a high risk of confusing disease symptoms of Lectera colletotrichoides with those caused by Ascochyta rabiei, the economic impact of Lectera colletotrichoides may be underestimated. It may be sensitive to agricultural pesticides, but is unlikely to face major threats from specific eradication measures. Its conservation status (
Africa: DRC: Mulungu, causing leaf spot of Glycine soja, comm. 28 Mar. 1978, D.J. Allen (IMI 226829a). Egypt: Ismailia, Salheia, isol. ex Cicer arietinum, 1988, M. Askar CP2035 (IMI 332702). Ethiopia: unlocalized, isol. ex seed of Phaseolus vulgaris, undated, s. coll. (IMI 366179). Morocco, unlocalized, isol. ex Capsicum annuum, Feb. 1986, Hasmi (IMI 303685). Nigeria: unlocalized, isol. ex Vigna sinensis, comm. 22 May 1972, R. Williams 4150 (IMI 166385); same data, R. Williams 4152 (IMI 166394). Asia: India: Rajupur, Mirzapur, on and isol. ex Senna sophera, Apr. 1982, S.N.P. Chaurasia SNPC-5 (IMI 269185); Jabalpur, isol. ex leaf litter of Tectona grandis, comm. 5 Feb. 1986, Jamaluddin (IMI 265505). Australasia: Australia: New South Wales, unlocalized, isol. ex Agrostis stolonifera, comm. 26 Jan. 1983, G. Schultz (IMI 275263); Western Australia: Esperance, isol. ex Trifolium subterraneum, comm. 13 Jul. 1972, R.F. Doepel (IMI 167533); New Zealand: Wellington, isol. ex Hordeum vulgare, comm. 11 Apr. 1980, L. Chong 18P14 (IMI 247318). Europe: United Kingdom: Kent, nr Ashford, Wye College, comm. 24 Oct. 1979, D.W. Parry 6 (IMI 242382); same locality, isol. ex Zea mays, comm. 24 Oct. 1979, D.W. Parry 11 (IMI 242387). South America: Argentina: Córdoba, San Francisco, Monte Cristo, isol. ex stem of Xanthium spinosum, 1995, s. coll. (IMI 368065). Brazil: unlocalized, isol. ex soil, comm. 18 Feb. 1982, J. Diehl & E. Reis 46/81 (IMI 265740).
Differs from Lectera colletotrichoides by its longer conidia (7.8–10 × 2–2.5 µm; mean 8.87 µm, sd 0.56, n=20) with a mean length/breadth ratio of 3.98: 1, with three short insertions and a single substitution in the ITS sequence.
Western Australia: Nedlands, isol. ex Triticum sp., 25 Jan. 1974, K. Sivasithamparam 530 (IMI 181698) – holotype of Lectera longa (dried specimen) and associated living culture.
Associated with Triticum sp. (Poaceae).
Only definitely known from the type locality.
No details are known; the data associated with the type do not indicate whether the fungus was thought to be pathogenic.
The species is only known from a single collection made in 1974, though a strain isolated from Viola sp. and identified as Volutella colletotrichoides with an identical ITS sequence (AJ301962) was deposited in the BBLF culture collection as BBA 71246. Its geographical origin is unknown, and the status of the living culture is uncertain. Its conservation status (
The genus Volutella is poorly researched and no modern monograph is available, but some of the more well-known species, including the type Volutella ciliata, were included in a recent phylogenetic study of Fusarium-like fungi (
Lectera has brightly coloured sporodochia surrounded by brown setae (those in true Volutella species are hyaline). Other sporodochial genera with these characteristics include Kutilakesa, Sarcopodium and Actinostilbe (
One other species of Volutella has orange sporodochia surrounded by brown tapering setae, in common with Lectera colletotrichoides, Volutella melaloma Berk. & Br. which was described from dead leaves of Carex in the UK (
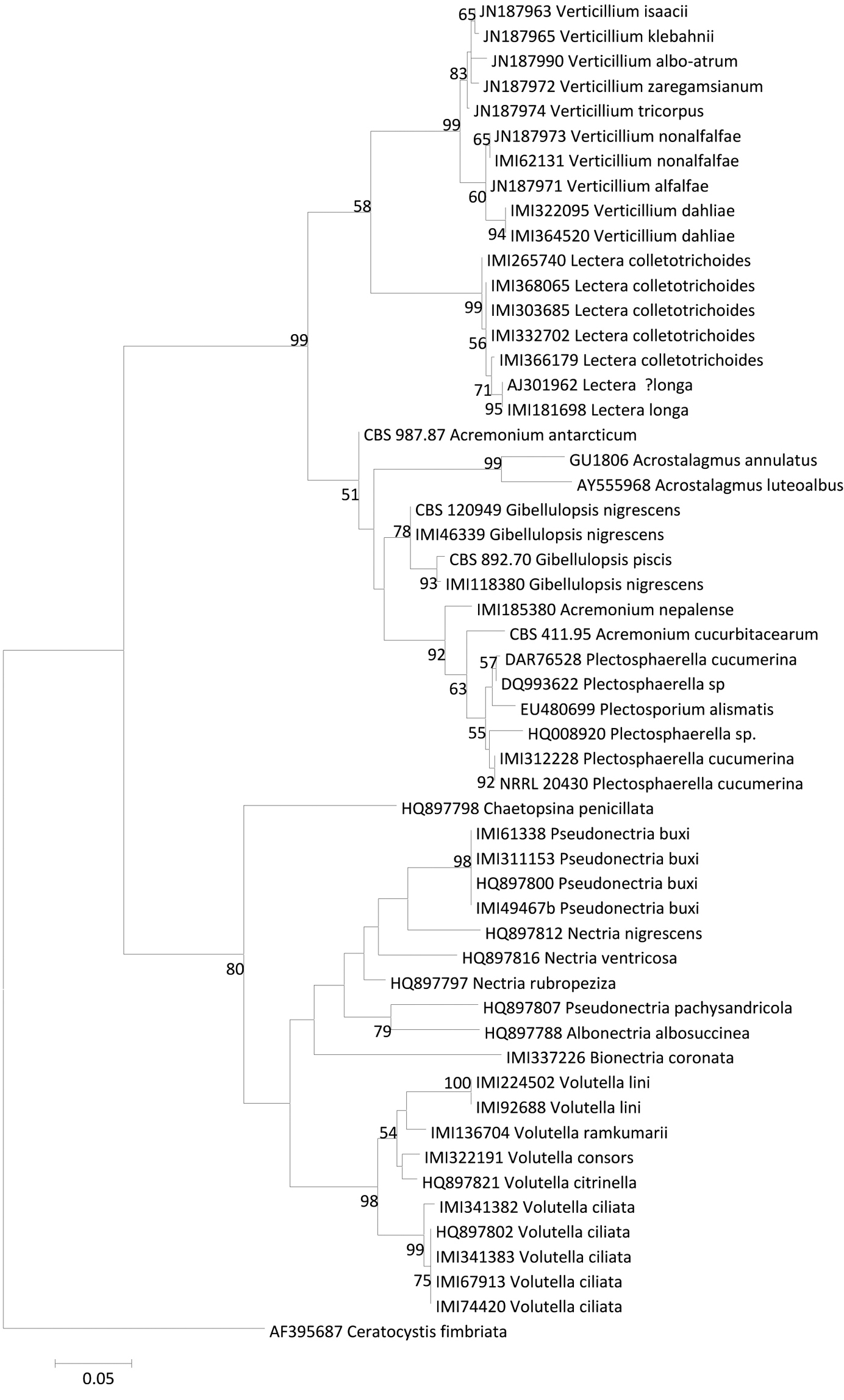
The ITS sequences generated in this study strongly suggest that Lectera has affinities with the Plectosphaerellaceae, and may be a sister group to Verticillium (Fig. 1), although this relationship only receives weak bootstrap support. That systematic relationship was previously noted incidentally by
Lectera colletotrichoides contains two distinct ITS phylotypes, IMI 265740 differing from the other strains sequenced by only a single base pair. The ITS sequence of Lectera longa differs from Lectera colletotrichoides by three short insertions and a single substitution, and this combined with differences in conidial length and lengh/width ratio justifies its separation. The distinction between the two species is supported by GAPDH sequences also (see fig. 3). The gene fragment sequenced here is a ~200 bp intron in the glyceraldehyde 3-phosphate dehydrogenase gene, used for phylogenetic studies of Colletotrichum by
ML ITS phyogram showing the phylogenetic position of Lectera species.
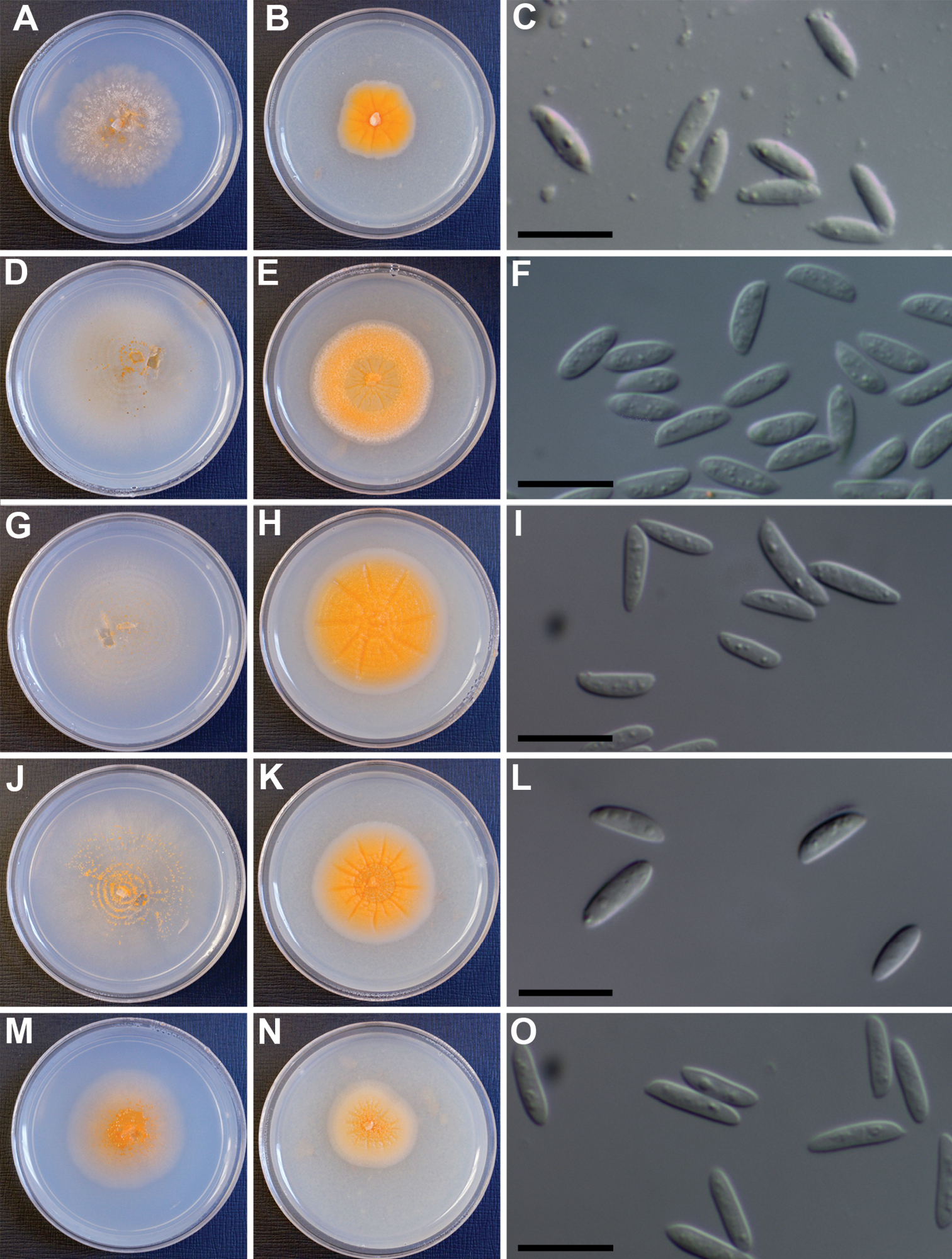
Lectera species. A–L Lectera colletotrichoides A–C IMI 166394 D–F, IMI 265740 G–I IMI 303685 J–L IMI 368065 M–O Lectera longa (IMI 181698) A, D, G, J, M colonies on PCA agar after 7 d B, E, H, K, N colonies on PDA agar after 7 d C, F, I, L, O: conidia mounted in lactic acid: bar = 10 µm.
ML tree of GAPDH sequences, of a subset of the strains used for the ITS sequence set.
The wide distribution of Lectera colletotrichoides and its association with agricultural ecosystems make it difficult to assess its geographical origin, although its apparent preference for dry-land legumes might indicate an evolutionary history centred on the Near East. However, judging from environmental sequencing studies, it (or a closely related species) does seem to be present in soils in SE USA (Wu et al. 1997,
We would like to thank Deborah Lewis, Curator of the Ada Hayden Herbarium, Iowa State University, for loan of type material of Volutella colletotrichoides, and for her generous donation of isolectotypes to the Kew Fungarium. In addition, we wish to acknowledge Jeniffer Lopez Castano, Saifuddien Haji Bagol, Zainab Kazaly and Lukasz Tymo for their work in sequencing some of the strains used in this study, and Helen Stewart for assistance with the culture work. The paper would not have been possible to prepare without the resources of CABI’s Genetic Resource collection.