|
||
|
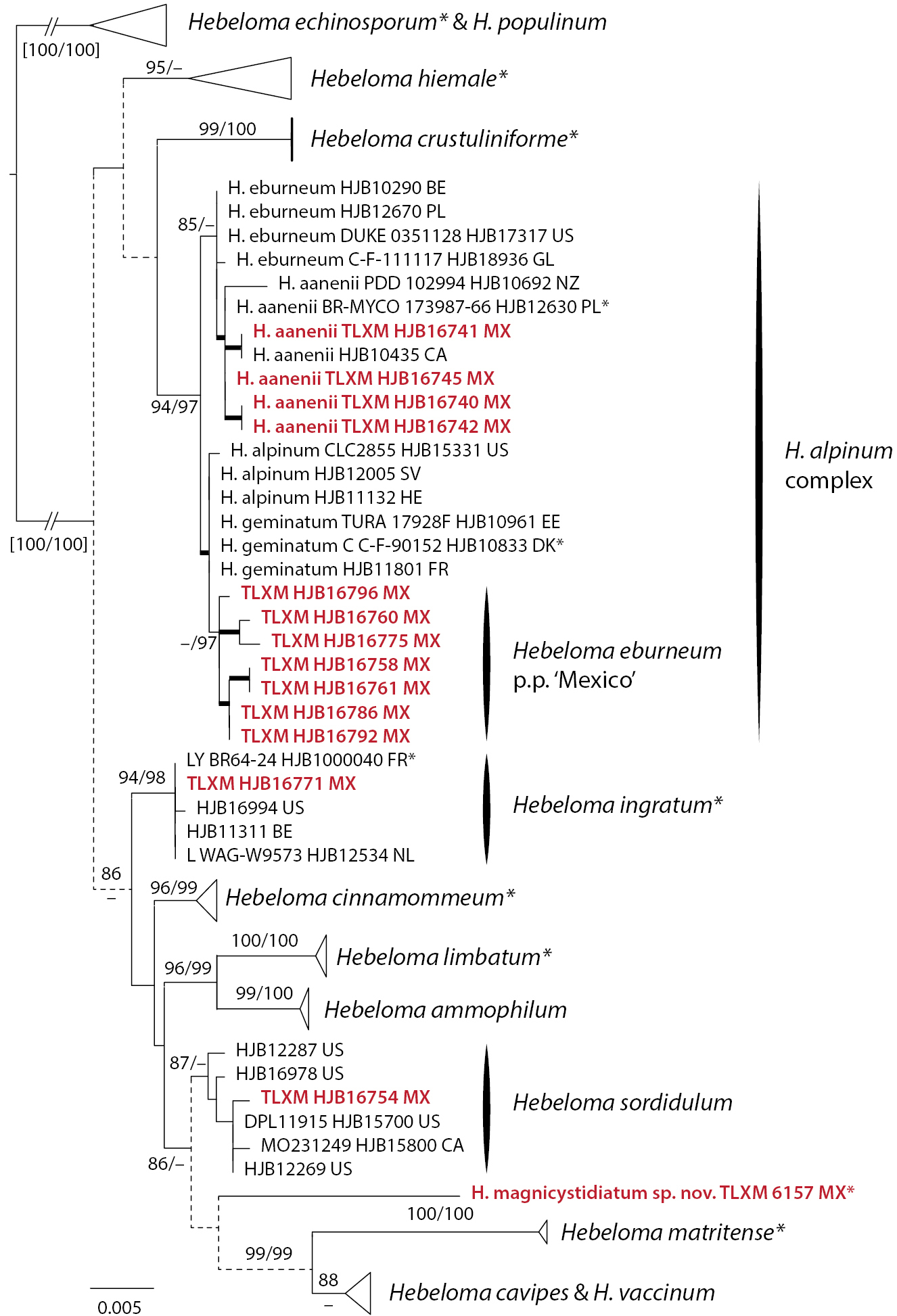
ML topology of concatenated ITS, mitSSU V6 and V9 sequences of Hebeloma sect. Denudata. Branch support was obtained through 1000 replicates of SH-like approximate likelihood ratio tests and ultrafast bootstrap annotated SH-aLRT/ufb at the branches for ≥ 85% SH-aLRT and ≥ 95% for ufb support or by thick lines in the case that at least one of the support values is equal to or exceeds the limits. Dotted lines indicate parts of the tree where conflicts between single locus results were observed. Hebeloma echinosporum and H. populinum (H. subsect. Echinospora of H. sect. Denudata) were used for rooting. Collections indicated with * are types; clade names indicated by * include type sequences. Collections in red refer to Mexican material. |