|
||
|
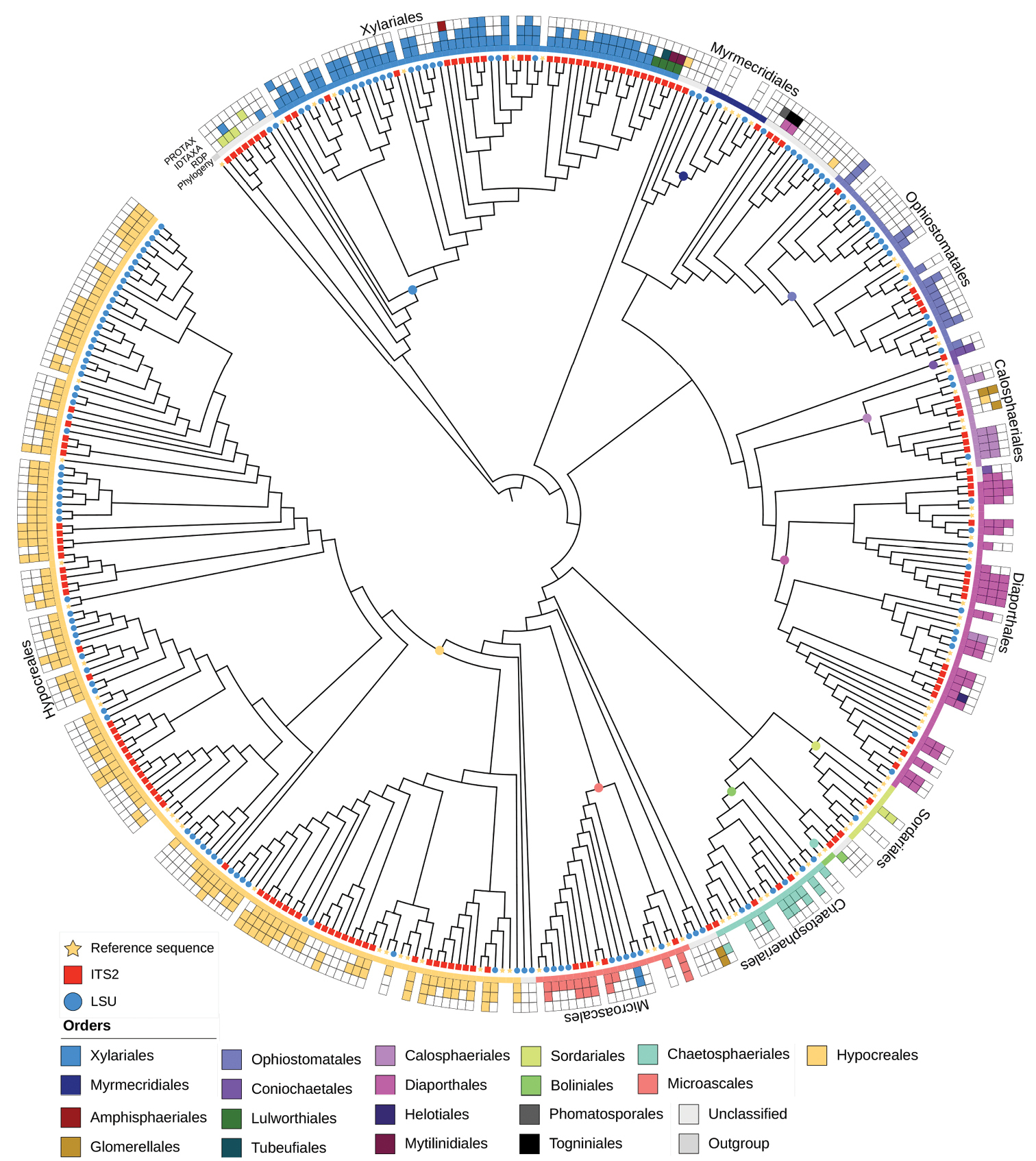
ML tree of Sordariomycetes constructed from the reference sequence alignments and OTUs for both markers (clustering thresholds: 98% ITS2, 99% LSU D1-D2). Leotia lubrica (Leotiomycetes) was specified as the outgroup. The assignment of OTUs by each of the three classifiers (RDP, IDTAXA, Protax-fungi) is shown by coloured boxes. Terminals missing these boxes are the reference sequences. Coloured dots on the nodes of the tree indicate the hypothetical ancestor defining monophyletic groups corresponding to the various orders of Sordariomycetes. The extent of each order is indicated by the coloured inner ring. Note that the ancestor of an order is defined by the youngest node from which all reference sequences are descended; OTUs falling outside of the resulting clades appear as ‘unassigned’ by the phylogenetic analysis approach. The distribution of ITS2 (red squares) and LSU D1-D2 (blue bullets) relative to the reference set (yellow stars) on each of the tips of the tree. Note the limited presence of ITS sequences in the Ophiostomatales (in top right quadrant). |