|
||
|
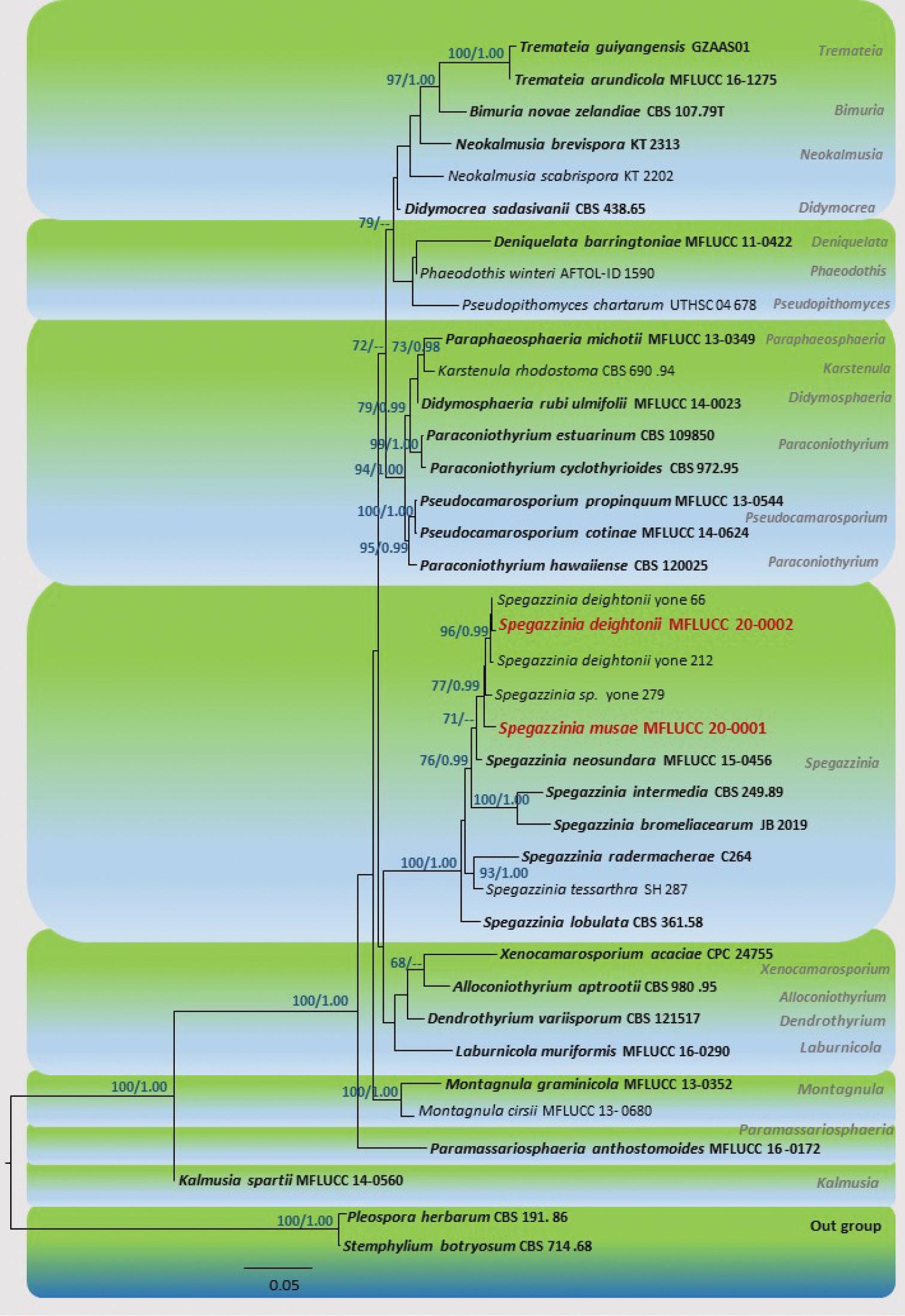
Maximum likelihood tree revealed by RAxML from an analysis of SSU, LSU and ITS and TEF1-α sequence data of selected genera of family Didymosphaeriaceae, showing the phylogenetic position of Spegazzinia musae (MFLUCC 20-0001) and S. deightonii (MFLUCC 20-0002). ML bootstrap supports (≥60 %) and Bayesian posterior probabilities (≥ 0.95 BYPP) are given above in the branches, respectively. The tree was rooted with Pleospora herbarum and Stemphylium botryosum (Pleosporaceae). Strains generated in this study are indicated in red-bold. Ex-type species are indicated in bold. The scale bar represents the expected number of nucleotide substitutions per site. A best scoring RAxML tree is shown with a final ML optimization likelihood value of -13516.66. The matrix had 795 distinct alignment patterns, with 33.60% of undetermined characters or gaps. Estimated base frequencies were: A = 0.239862, C = 0.245185, G = 0.277025, T = 0.237927; substitution rates AC = 1.626982, AG = 2.468452, AT = 1.211822, CG = 1.092437, CT = 6.295657, GT = 1.000000; proportion of invariable sites I = 0.484119; gamma distribution shape parameter α = 0.445929. |