|
||
|
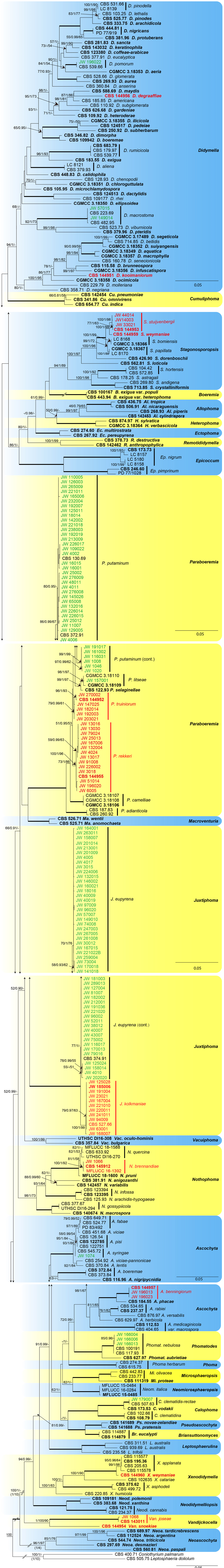
Phylogenetic tree generated from the maximum-likelihood analysis based on the combined ITS, LSU, tub2 and rpb2 sequence alignment of Didymellaceae members. The RAxML bootstrap support values (BS), Bayesian posterior probabilities (PP), and parsimony bootstrap support values (PBS) are given at the nodes (BS/PP/PBS). BS and PBS values represent parsimony bootstrap support values >50 %. Full supported branches are indicated in bold. The scale bar represents the expected number of changes per site. Ex-type strains are represented in bold. Strains obtained in the current study are printed in green; among them, whilst strains that represent new taxa are printed in red. Some of the basal branches were shortened to facilitate layout (the fraction in round parentheses refers to the presented length compared to the actual length of the branch). The tree was rooted to Coniothyrium palmarum CBS 400.71 and Leptosphaeria doliolum CBS 505.75. |