|
||
|
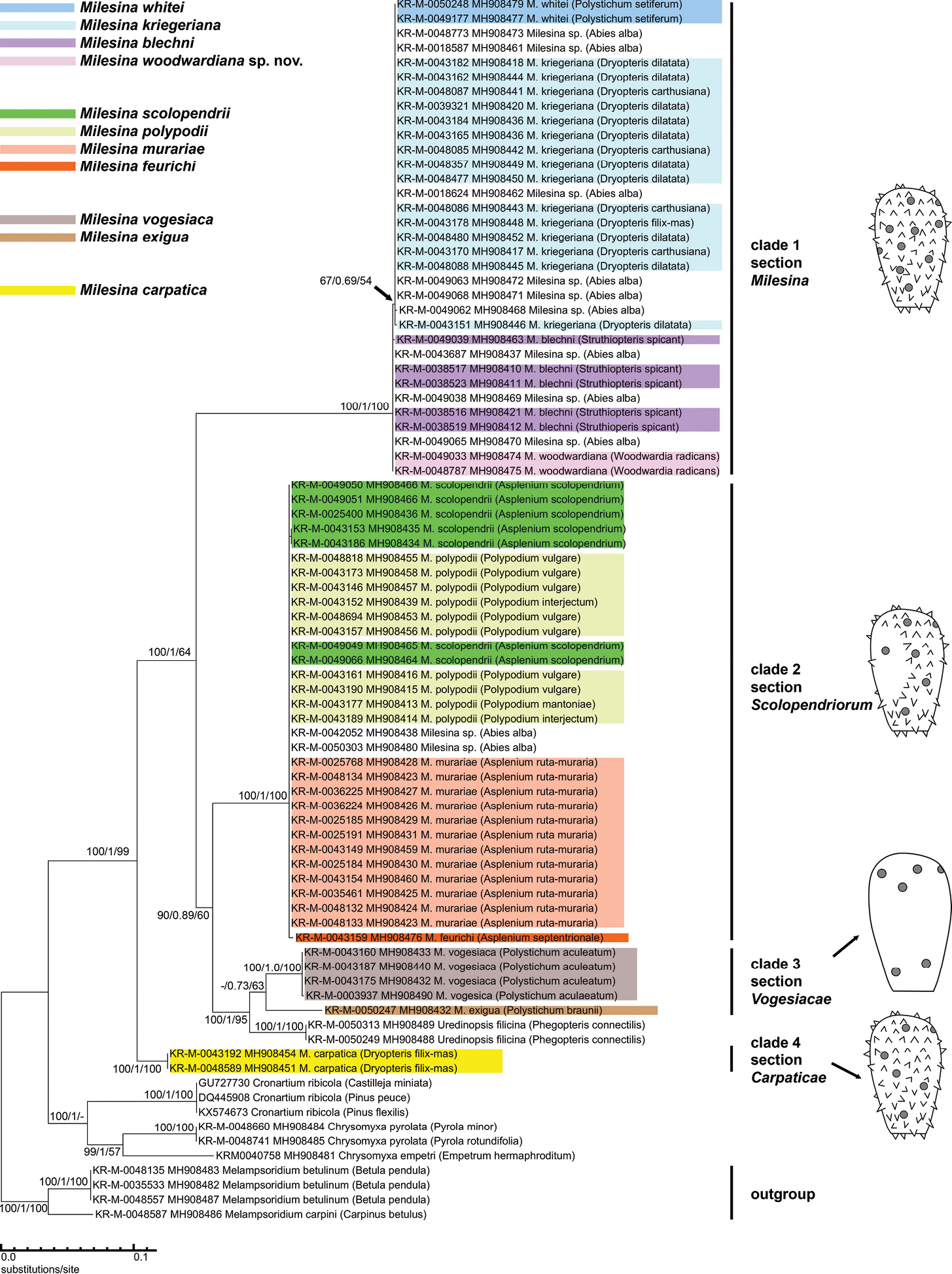
ITS Phylogram of 11 Milesina species (excluding M. magnusiana). The phylogram is based on a 733-bp alignment. A Maximum Likelihood (ML) tree is shown with support values for ML, Bayesian Inference (BI) and Neighbour Joining (NJ), in the order ML/BI/NJ. Support values are presented when they are above 50 (ML, NJ) or 0.5 (BI). The host is indicated in brackets. Milesina specimens without species designation (host Abies alba) are not colour-coded. For comparison, several sequences were included from closely related genera. They were all newly generated within the GBOL project, except the GenBank sequences for Cronartium spp. The drawings on the right side present the typical arrangement of spines and germ pores (grey dots) on the Milesina urediniospores. |