|
||
|
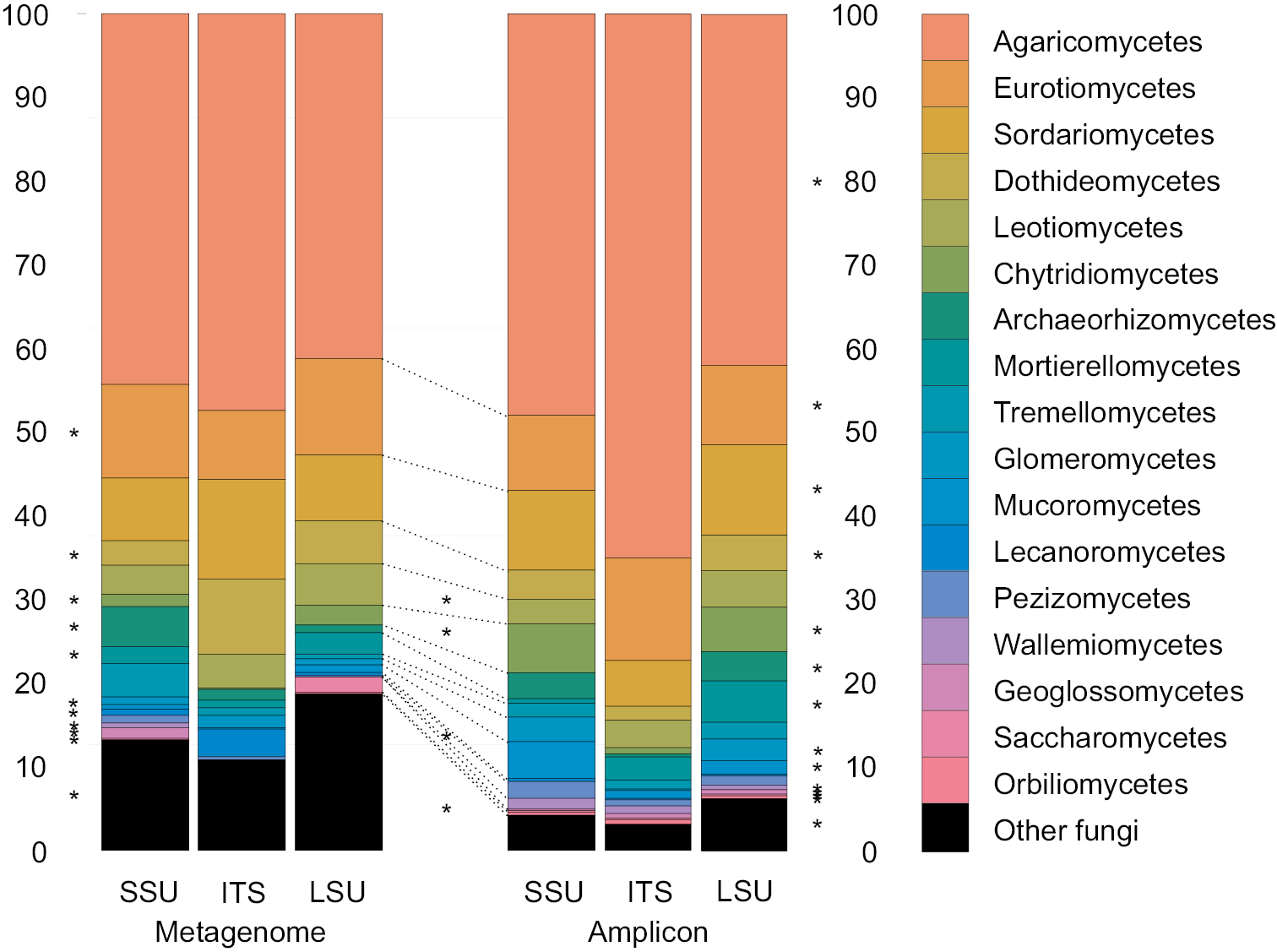
Relative abundance of fungal classes in the amplicon and metagenomics data sets divided into SSU, ITS, and LSU subsets averaged over different barcodes (amplicon data) and 14 shared samples. Asterisks in the margins indicate significant differences in recovery of classes among SSU, ITS, and LSU of metagenomics (right) and amplicon (left) data sets. Asterisks in the center indicate significant differences between the metagenomics and amplicon-bases approaches. |