|
||
|
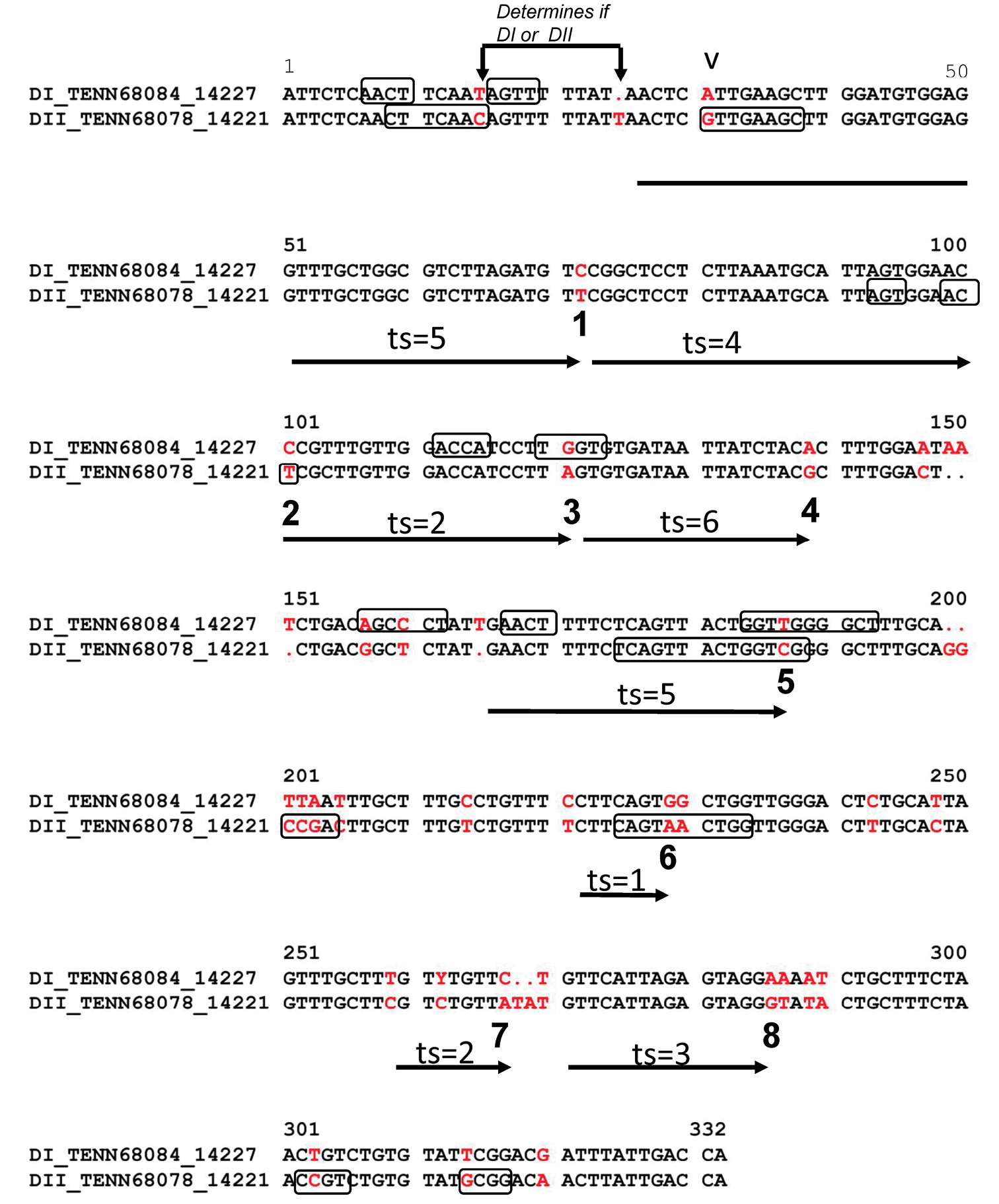
The ITS2 region of two haplotypes of Gymnopus dichrous: Haplotypes DI (represented by TENN68084) and DII (represented by TENN68078). The TC pair at position 15 and the indel at position 25 were used to determine which the haplotype was represented by the 5’ end of a cloned sequence. Bases in red are points where DI and DII haplotypes differ in sequence and were used to determine if template switching had occurred in a cloned PCR product. Eight base pairs at which template switching can be detected are indicated by numbers 1-8. The possible area in which template switching (ts) could have occurred is indicated by vertical arrows and the number of observed template switching events is given above the vertical arrow. Bases that may be involved in intra-strand base pairing as determined by MFOLD are outlined with black boxes. Ambiguity codes indicate intraspecific variation. |